I have stumbled across a problem in R that I was hoping someone could clear up why it is happening and how to fix it. I am not well vetted in the use of R and sometimes get muddled in the way that one line of code can often do much more than many other languages. The problem seems to be that the program isn't correctly taking the file inputs after the first. If I input one file, the histogram comes out the way I would expect. But unfortunately when more than one file is input it combines them and smooshes them next to the first. I would rather each input file have its own stand alone histogram. Sorry for the long post but I am trying to give as much info as I can to make my code reproducible (I am bad at reproducible code it seems).
The code is as such:
library("tcltk")
#choose any number of files
File.names<-(tk_choose.files(default="", caption="Choose your files", multi=TRUE, filters=NULL, index=1))
Num.Files<-NROW(File.names)
#read the tables
dat <- lapply(File.names,read.table,header = TRUE)
names(dat) <- paste("f", 1:length(Num.Files), sep="")
#use the 14th columns data
tmp <- stack(lapply(dat,function(x) x[,14]))
#this is where the histogram is made(with percent shown on the y axis)
require(ggplot2)
ggplot(tmp,aes(x = values)) +
facet_wrap(~ind) +
geom_histogram(aes(y=..count../sum(..count..)))
dput(tmp)
dput(dat)
sessionInfo()
Here is an example of a file that could be chosen by the user:
Targ cov av_cov 87A_cvg 87Ag 87Agr 87Agr 87A_gra 87A%_1 87A%_3 87A%_5 87A%_10 87A%_20 87A%_30 87A%_40 87A%_50 87A%_75 87A%_100
1:028 400 0.42 400 0.42 1 1 2 41.8 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
1:296 400 0.42 400 0.42 1 1 2 41.8 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
1:453 1646 8.11 1646 8.11 7 8 13 100.0 100.0 87.2 32.0 0.0 0.0 0.0 0.0 0.0 0.0
1:427 1646 8.11 1646 8.11 7 8 13 100.0 100.0 87.2 32.0 0.0 0.0 0.0 0.0 0.0 0.0
1:736 5105 29.68 5105 29.68 14 29 48 100.0 100.0 100.0 86.0 65.7 49.4 35.5 16.9 0.0 0.0
1:514 5105 29.68 5105 29.68 14 29 48 100.0 100.0 100.0 86.0 65.7 49.4 35.5 16.9 0.0 0.0
1:296 5105 29.68 5105 29.68 14 29 48 100.0 100.0 100.0 86.0 65.7 49.4 35.5 16.9 0.0 0.0
1:534 400 0.42 400 0.42 1 1 2 41.8 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0
And another:
Targ cov av_cov 87A_cvg 87Ag 87Agr 87Agr 87A_gra 87A%_1 87A%_3 87A%_5 87A%_10 87A%_20 87A%_30 87A%_40 87A%_50 87A%_75 87A%_100
1:028 400 0.42 400 0.42 1 1 2 41.8 0.0 1.0 0.0 20.0 0.0 0.0 0.0 0.0 0.0
1:296 400 0.42 400 0.42 1 1 2 41.8 0.0 20.0 0.0 40.0 0.0 100.0 10.0 50.0 4.0
1:453 1646 8.11 1646 8.11 7 8 13 100.0 100.0 87.2 32.0 0.0 100.0 4.0 60.0 30.0 20.0
1:427 1646 8.11 1646 8.11 7 8 13 100.0 100.0 87.2 32.0 0.0 80.0 40.0 60.0 80.0 90.0
1:736 5105 29.68 5105 29.68 14 29 48 100.0 100.0 100.0 86.0 65.7 49.4 35.5 16.9 30.0 20.0
1:514 5105 29.68 5105 29.68 14 29 48 100.0 100.0 100.0 86.0 65.7 49.4 35.5 16.9 20.0 30.0
1:296 5105 29.68 5105 29.68 14 29 48 100.0 100.0 100.0 86.0 65.7 49.4 35.5 16.9 20.0 30.0
1:534 400 0.42 400 0.42 1 1 2 41.8 0.0 40.0 30.0 80.0 70.0 40.0 30.0 30.0 10.0
The code works well with one file(these histograms are made from different input files but you get the picture) but disagrees with multiple files (regardless of the number):
One:
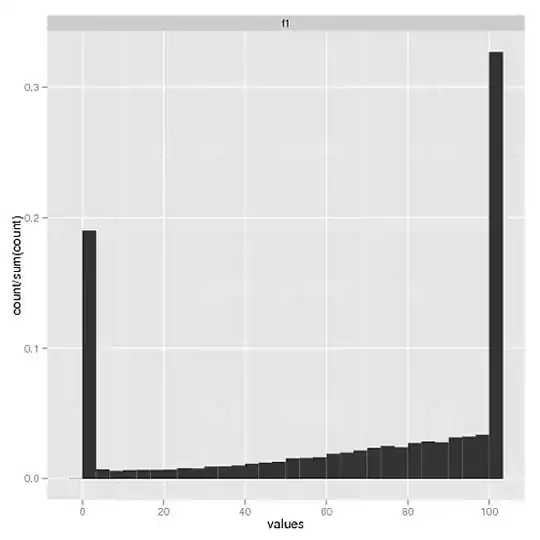
This is how I hope all of the histograms to look, one for each entered file. But alas...
Multiple files:
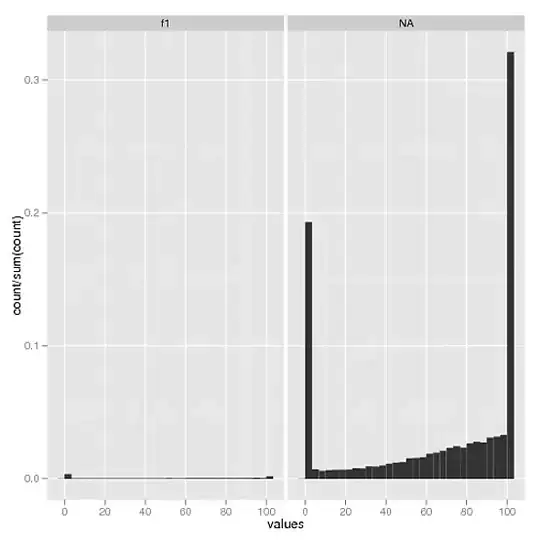
> dput(tmp)
structure(list(values = c(0, 0, 0, 0, 49.4, 49.4, 49.4, 0), ind = structure(c(1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L), .Label = "f1", class = "factor")), .Names = c("values",
"ind"), row.names = c(NA, -8L), class = "data.frame")
> dput(dat)
structure(list(f1 = structure(list(Targ = structure(c(1L, 2L,
4L, 3L, 7L, 5L, 2L, 6L), .Label = c("1:028", "1:296", "1:427",
"1:453", "1:514", "1:534", "1:736"), class = "factor"), cov = c(400L,
400L, 1646L, 1646L, 5105L, 5105L, 5105L, 400L), av_cov = c(0.42,
0.42, 8.11, 8.11, 29.68, 29.68, 29.68, 0.42), "X87A_cvg", "X87Ag", "X87Agr", "X87Agr.1", "X87A_gra", "X87A._1", "X87A._3", "X87A._5", "X87A._10", "X87A._20", "X87A._30", "X87A._40",
"X87A._50", "X87A._75", "X87A._100"), class = "data.frame", row.names = c(NA,
-8L))), .Names = "f1")
> sessionInfo()
R version 2.14.1 (2011-12-22)
Platform: x86_64-redhat-linux-gnu (64-bit)
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=C LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] tcltk stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] ggplot2_0.9.1
loaded via a namespace (and not attached):
[1] colorspace_1.1-1 dichromat_1.2-4 digest_0.5.2 grid_2.14.1
[5] labeling_0.1 MASS_7.3-17 memoise_0.1 munsell_0.3
[9] plyr_1.7.1 proto_0.3-9.2 RColorBrewer_1.0-5 reshape2_1.2.1
[13] scales_0.2.1 stringr_0.6
Is there any way to make each histogram separate, and able to stand alone? Thanks in advance Steph