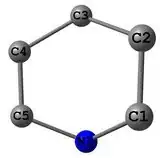
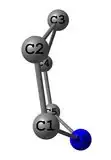
I have 4 points, which are very near to be at the one plane - it is the 1,4-Dihydropyridine cycle.
I need to calculate distance from C3 and N1 to the plane, which is made of C1-C2-C4-C5. Calculating distance is OK, but fitting plane is quite difficult to me.
1,4-DHP cycle:
1,4-DHP cycle, another view:
from array import *
from numpy import *
from scipy import *
# coordinates (XYZ) of C1, C2, C4 and C5
x = [0.274791784, -1.001679346, -1.851320839, 0.365840754]
y = [-1.155674199, -1.215133985, 0.053119249, 1.162878076]
z = [1.216239624, 0.764265677, 0.956099579, 1.198231236]
# plane equation Ax + By + Cz = D
# non-fitted plane
abcd = [0.506645455682, -0.185724560275, -1.43998120646, 1.37626378129]
# creating distance variable
distance = zeros(4, float)
# calculating distance from point to plane
for i in range(4):
distance[i] = (x[i]*abcd[0]+y[i]*abcd[1]+z[i]*abcd[2]+abcd[3])/sqrt(abcd[0]**2 + abcd[1]**2 + abcd[2]**2)
print distance
# calculating squares
squares = distance**2
print squares
How to make sum(squares) minimized? I have tried least squares, but it is too hard for me.