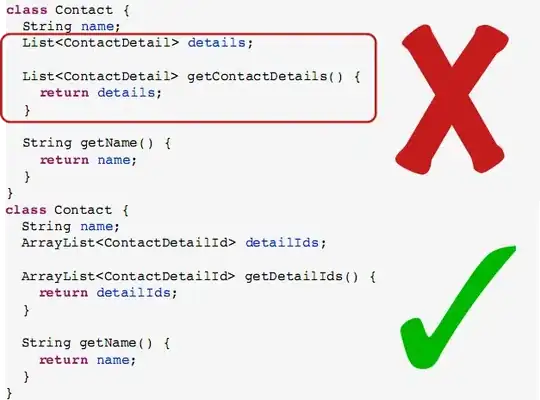
I created a rose diagram of aspects in degrees for location data using the 'circular' package in R and the rose.diag function, with basic aspects of N, NE, E, etc., for a total of 8 bins. However, the bins do not straddle the aspects. In other words, the first bin goes from 0-45, the 2nd from 45 to 90, and so on, which is pooling the aspect data in strange ways. Is there any way to shift the bins so 0, 45, 90, etc are the center of the bins, instead of the edges?
rose.diag(Degrees$Degrees, bins=8,zero=pi/2, units = 'degrees', rotation='clock')
 This may not be ideal because not all of the features in
This may not be ideal because not all of the features in