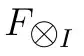Is there some way to use rollapply (from zoo package or something similar) optimized functions (rollmean, rollmedian etc) to compute rolling functions with a time-based window, instead of one based on a number of observations? What I want is simple: for each element in an irregular time series, I want to compute a rolling function with a N-days window. That is, the window should include all the observations up to N days before the current observation. Time series may also contain duplicates.
Here follows an example. Given the following time series:
date value
1/11/2011 5
1/11/2011 4
1/11/2011 2
8/11/2011 1
13/11/2011 0
14/11/2011 0
15/11/2011 0
18/11/2011 1
21/11/2011 4
5/12/2011 3
A rolling median with a 5-day window, aligned to the right, should result in the following calculation:
> c(
median(c(5)),
median(c(5,4)),
median(c(5,4,2)),
median(c(1)),
median(c(1,0)),
median(c(0,0)),
median(c(0,0,0)),
median(c(0,0,0,1)),
median(c(1,4)),
median(c(3))
)
[1] 5.0 4.5 4.0 1.0 0.5 0.0 0.0 0.0 2.5 3.0
I already found some solutions out there but they are usually tricky, which usually means slow. I managed to implement my own rolling function calculation. The problem is that for very long time series the optimized version of median (rollmedian) can make a huge time difference, since it takes into account the overlap between windows. I would like to avoid reimplementing it. I suspect there are some trick with rollapply parameters that will make it work, but I cannot figure it out. Thanks in advance for the help.