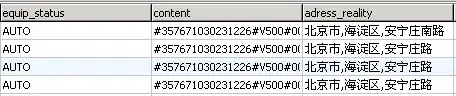
I had similar problem and google pointed me to this: blog post. Basically he's using inpaint algorithm to interpolate missing values and produce valid array for filtering.
The code is at the end of the post, and you can save it to site-packages (or else) and load it as module (i.e. inpaint.py):
import inpaint
filled = inpaint.replace_nans(NANMask, 5, 0.5, 2, 'idw')
I'm happy with the result, and I guess it will suite missing temperature values just fine. There is also next version here: github but code will need some cleaning for general usage as it's part of a project.
For reference, easy use and preservation sake I'll post the code (of initial version) here:
# -*- coding: utf-8 -*-
"""A module for various utilities and helper functions"""
import numpy as np
#cimport numpy as np
#cimport cython
DTYPEf = np.float64
#ctypedef np.float64_t DTYPEf_t
DTYPEi = np.int32
#ctypedef np.int32_t DTYPEi_t
#@cython.boundscheck(False) # turn of bounds-checking for entire function
#@cython.wraparound(False) # turn of bounds-checking for entire function
def replace_nans(array, max_iter, tol,kernel_size=1,method='localmean'):
"""Replace NaN elements in an array using an iterative image inpainting algorithm.
The algorithm is the following:
1) For each element in the input array, replace it by a weighted average
of the neighbouring elements which are not NaN themselves. The weights depends
of the method type. If ``method=localmean`` weight are equal to 1/( (2*kernel_size+1)**2 -1 )
2) Several iterations are needed if there are adjacent NaN elements.
If this is the case, information is "spread" from the edges of the missing
regions iteratively, until the variation is below a certain threshold.
Parameters
----------
array : 2d np.ndarray
an array containing NaN elements that have to be replaced
max_iter : int
the number of iterations
kernel_size : int
the size of the kernel, default is 1
method : str
the method used to replace invalid values. Valid options are
`localmean`, 'idw'.
Returns
-------
filled : 2d np.ndarray
a copy of the input array, where NaN elements have been replaced.
"""
# cdef int i, j, I, J, it, n, k, l
# cdef int n_invalids
filled = np.empty( [array.shape[0], array.shape[1]], dtype=DTYPEf)
kernel = np.empty( (2*kernel_size+1, 2*kernel_size+1), dtype=DTYPEf )
# cdef np.ndarray[np.int_t, ndim=1] inans
# cdef np.ndarray[np.int_t, ndim=1] jnans
# indices where array is NaN
inans, jnans = np.nonzero( np.isnan(array) )
# number of NaN elements
n_nans = len(inans)
# arrays which contain replaced values to check for convergence
replaced_new = np.zeros( n_nans, dtype=DTYPEf)
replaced_old = np.zeros( n_nans, dtype=DTYPEf)
# depending on kernel type, fill kernel array
if method == 'localmean':
print 'kernel_size', kernel_size
for i in range(2*kernel_size+1):
for j in range(2*kernel_size+1):
kernel[i,j] = 1
print kernel, 'kernel'
elif method == 'idw':
kernel = np.array([[0, 0.5, 0.5, 0.5,0],
[0.5,0.75,0.75,0.75,0.5],
[0.5,0.75,1,0.75,0.5],
[0.5,0.75,0.75,0.5,1],
[0, 0.5, 0.5 ,0.5 ,0]])
print kernel, 'kernel'
else:
raise ValueError( 'method not valid. Should be one of `localmean`.')
# fill new array with input elements
for i in range(array.shape[0]):
for j in range(array.shape[1]):
filled[i,j] = array[i,j]
# make several passes
# until we reach convergence
for it in range(max_iter):
print 'iteration', it
# for each NaN element
for k in range(n_nans):
i = inans[k]
j = jnans[k]
# initialize to zero
filled[i,j] = 0.0
n = 0
# loop over the kernel
for I in range(2*kernel_size+1):
for J in range(2*kernel_size+1):
# if we are not out of the boundaries
if i+I-kernel_size < array.shape[0] and i+I-kernel_size >= 0:
if j+J-kernel_size < array.shape[1] and j+J-kernel_size >= 0:
# if the neighbour element is not NaN itself.
if filled[i+I-kernel_size, j+J-kernel_size] == filled[i+I-kernel_size, j+J-kernel_size] :
# do not sum itself
if I-kernel_size != 0 and J-kernel_size != 0:
# convolve kernel with original array
filled[i,j] = filled[i,j] + filled[i+I-kernel_size, j+J-kernel_size]*kernel[I, J]
n = n + 1*kernel[I,J]
# divide value by effective number of added elements
if n != 0:
filled[i,j] = filled[i,j] / n
replaced_new[k] = filled[i,j]
else:
filled[i,j] = np.nan
# check if mean square difference between values of replaced
#elements is below a certain tolerance
print 'tolerance', np.mean( (replaced_new-replaced_old)**2 )
if np.mean( (replaced_new-replaced_old)**2 ) < tol:
break
else:
for l in range(n_nans):
replaced_old[l] = replaced_new[l]
return filled
def sincinterp(image, x, y, kernel_size=3 ):
"""Re-sample an image at intermediate positions between pixels.
This function uses a cardinal interpolation formula which limits
the loss of information in the resampling process. It uses a limited
number of neighbouring pixels.
The new image :math:`im^+` at fractional locations :math:`x` and :math:`y` is computed as:
.. math::
im^+(x,y) = \sum_{i=-\mathtt{kernel\_size}}^{i=\mathtt{kernel\_size}} \sum_{j=-\mathtt{kernel\_size}}^{j=\mathtt{kernel\_size}} \mathtt{image}(i,j) sin[\pi(i-\mathtt{x})] sin[\pi(j-\mathtt{y})] / \pi(i-\mathtt{x}) / \pi(j-\mathtt{y})
Parameters
----------
image : np.ndarray, dtype np.int32
the image array.
x : two dimensions np.ndarray of floats
an array containing fractional pixel row
positions at which to interpolate the image
y : two dimensions np.ndarray of floats
an array containing fractional pixel column
positions at which to interpolate the image
kernel_size : int
interpolation is performed over a ``(2*kernel_size+1)*(2*kernel_size+1)``
submatrix in the neighbourhood of each interpolation point.
Returns
-------
im : np.ndarray, dtype np.float64
the interpolated value of ``image`` at the points specified
by ``x`` and ``y``
"""
# indices
# cdef int i, j, I, J
# the output array
r = np.zeros( [x.shape[0], x.shape[1]], dtype=DTYPEf)
# fast pi
pi = 3.1419
# for each point of the output array
for I in range(x.shape[0]):
for J in range(x.shape[1]):
#loop over all neighbouring grid points
for i in range( int(x[I,J])-kernel_size, int(x[I,J])+kernel_size+1 ):
for j in range( int(y[I,J])-kernel_size, int(y[I,J])+kernel_size+1 ):
# check that we are in the boundaries
if i >= 0 and i <= image.shape[0] and j >= 0 and j <= image.shape[1]:
if (i-x[I,J]) == 0.0 and (j-y[I,J]) == 0.0:
r[I,J] = r[I,J] + image[i,j]
elif (i-x[I,J]) == 0.0:
r[I,J] = r[I,J] + image[i,j] * np.sin( pi*(j-y[I,J]) )/( pi*(j-y[I,J]) )
elif (j-y[I,J]) == 0.0:
r[I,J] = r[I,J] + image[i,j] * np.sin( pi*(i-x[I,J]) )/( pi*(i-x[I,J]) )
else:
r[I,J] = r[I,J] + image[i,j] * np.sin( pi*(i-x[I,J]) )*np.sin( pi*(j-y[I,J]) )/( pi*pi*(i-x[I,J])*(j-y[I,J]))
return r
#cdef extern from "math.h":
# double sin(double)