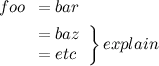I did a linear discriminant analysis using the function lda() from the package MASS. Now I would try to plot a biplot like in ade4 package (forLDA). Do you know how can I do this?
If I try to use the biplot() function it doesn't work. For example, if I use the Iris data and make LDA:
dis2 <- lda(as.matrix(iris[, 1:4]), iris$Species)
then I can plot it using the function plot(), but if I use the function biplot() it doesn't work:
biplot(dis2)
Error in nrow(y) : argument "y" is missing, with no default
How can I plot the arrows of variables?