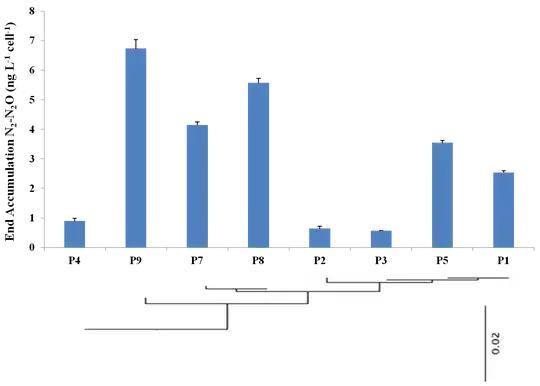
I'm trying to arrange a phylogenetic tree onto a graph showing physiological data for a set of related organisms. Something like the picture below. This was put together in powerpoint from 2 separate graphs. I guess it gets the job done, but I was hoping to create a single image which I think will be easier to format into a document. I am able to produce the graph I want using ggplot2, and import the tree using ape. I was thinking there should be a way to save the tree as a graphical object and then arrange it with the graph using the gridarrange function in gridExtra. The problem is that ape won't let me save the tree as a graphical object, e.g.,
p2<-plot(tree, dir = "u", show.tip.label = FALSE)
just plots the tree and when you call p2 it just gives a list of arguments. I'm wondering if anyone has any tips.
Thanks!