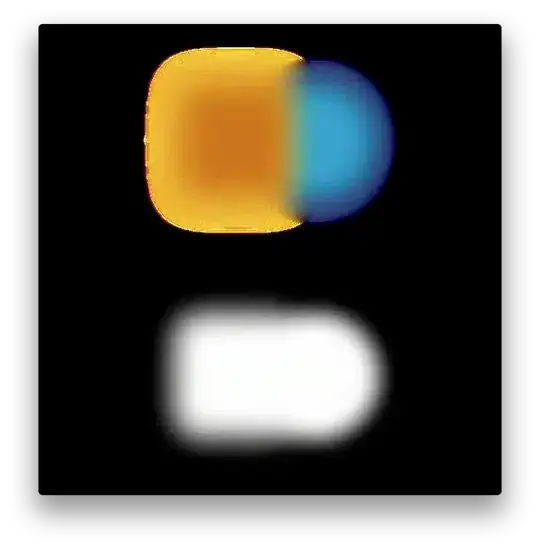
Hello! I have some points on a line. These points do not have an Y dimension, only an X dimension. I only placed them in an Y dimension because this wanted to be able to place multiple dots on the same spot.
I would like find n centroids (spots with the most density).
I placed for example centroids (=green lines) to show what I mean. These examplary centroids were not calculated, I only placed them guessing where they would be.
Before I dive into the math, I would like to know if this is can be solved with k-means-clustering, or if I am going in the wrong direction.
Thank you.