My function gets a combinations of numbers, can be small or large depending on user input, and will loop through each combinations to perform some operations.
Below is a line-profiling I ran on my function and it is takes 0.336 seconds to run. While this is fast, this is only a subset of a bigger framework. In this bigger framework, I will need to run this function 50-20000 times which when multiplied to 0.336, takes 16.8 to 6720 seconds (I hope this is right). Before it takes 0.996 seconds but I've manage to cut it by half through avoiding functions calls.
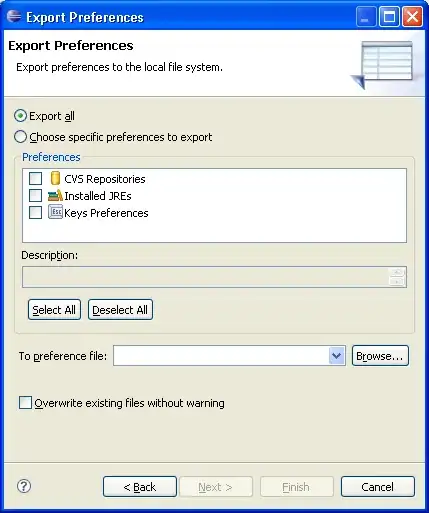
The major contributor to time is the two __getitem__ which is accessing dictionary for information N times depending on the number of combinations. My dictionary is a collection data and it looks something like this:
dic = {"array1", a,
"array2", b,
"array3", c,
"listofarray", [ [list of 5 array], [list of 5 array], [list of 5 2d Array ] ]
}
I was able to cut it by another ~0.01 seconds when I placed the dictionary lookback outside of the loop..
x = dic['listofarray']['list of 5 2d array']
So when I loop to get access to the the 5 different elements I just did x[i].
Other than that I am lost in terms of where to add more performance boost.
Note: I apologize that I haven't provided any code. I'd love to show but its proprietary. I just wanted to get some thoughts on whether I am looking at the right place for speed ups.
I am willing to learn and apply new things so if cython or some other data structure can speed things up, i am all ears. Thanks so much
PS:
inside my first __getitem__:

inside my second __getitem__:
EDIT:
I am using iter tools product(xrange(10), repeat=len(food_choices)) and iterating over this. I covert everything into numpy arrays np.array(i).astype(float).