I have a problem where I have a bunch of lengths and want to start at the origin (pretend I'm facing to the positive end of the y axis), I make a right and move positively along the x axis for the distance of length_i. At this time I make another right turn, walk the distance of length_i and repeat n times. I can do this but I think there's a more efficient way to do it and I lack a math background:
## Fake Data
set.seed(11)
dat <- data.frame(id = LETTERS[1:6], lens=sample(2:9, 6),
x1=NA, y1=NA, x2=NA, y2=NA)
## id lens x1 y1 x2 y2
## 1 A 4 NA NA NA NA
## 2 B 2 NA NA NA NA
## 3 C 5 NA NA NA NA
## 4 D 8 NA NA NA NA
## 5 E 6 NA NA NA NA
## 6 F 9 NA NA NA NA
## Add a cycle of 4 column
dat[, "cycle"] <- rep(1:4, ceiling(nrow(dat)/4))[1:nrow(dat)]
##For loop to use the information from cycle column
for(i in 1:nrow(dat)) {
## set x1, y1
if (i == 1) {
dat[1, c("x1", "y1")] <- 0
} else {
dat[i, c("x1", "y1")] <- dat[(i - 1), c("x2", "y2")]
}
col1 <- ifelse(dat[i, "cycle"] %% 2 == 0, "x1", "y1")
col2 <- ifelse(dat[i, "cycle"] %% 2 == 0, "x2", "y2")
dat[i, col2] <- dat[i, col1]
col3 <- ifelse(dat[i, "cycle"] %% 2 != 0, "x2", "y2")
col4 <- ifelse(dat[i, "cycle"] %% 2 != 0, "x1", "y1")
mag <- ifelse(dat[i, "cycle"] %in% c(1, 4), 1, -1)
dat[i, col3] <- dat[i, col4] + (dat[i, "lens"] * mag)
}
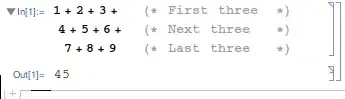
This gives the desired result:
> dat
id lens x1 y1 x2 y2 cycle
1 A 4 0 0 4 0 1
2 B 2 4 0 4 -2 2
3 C 5 4 -2 -1 -2 3
4 D 8 -1 -2 -1 6 4
5 E 6 -1 6 5 6 1
6 F 9 5 6 5 -3 2
Here it is as a plot:
library(ggplot2); library(grid)
ggplot(dat, aes(x = x1, y = y1, xend = x2, yend = y2)) +
geom_segment(aes(color=id), size=3, arrow = arrow(length = unit(0.5, "cm"))) +
ylim(c(-10, 10)) + xlim(c(-10, 10))
This seems slow and clunky. I'm guessing there's a better way to do this than the items I do in the for loop. What's a more efficient way to keep making programatic rights?