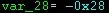I'll try to approach this by mosaic()'ing images with different extents and origins but same resolution.
Create a few rasterLayer objects and export them (to read latter)
library('raster')
library('rgdal')
e1 <- extent(0,10,0,10)
r1 <- raster(e1)
res(r1) <- 0.5
r1[] <- runif(400, min = 0, max = 1)
#plot(r1)
e2 <- extent(5,15,5,15)
r2 <- raster(e2)
res(r2) <- 0.5
r2[] <- rnorm(400, 5, 1)
#plot(r2)
e3 <- extent(18,40,18,40)
r3 <- raster(e3)
res(r3) <- 0.5
r3[] <- rnorm(1936, 12, 1)
#plot(r3)
# Write them out
wdata <- '../Stackoverflow/21876858' # your local folder
writeRaster(r1, file.path(wdata, 'r1.tif'),
overwrite = TRUE)
writeRaster(r2,file.path(wdata, 'r2.tif'),
overwrite = TRUE)
writeRaster(r3,file.path(wdata, 'r3.tif'),
overwrite = TRUE)
Read and Mosaic'ing with function
Since raster::mosaic do not accept rasterStack/rasterBrick or lists of rasterLayers, the best approach is to use do.call, like this excellent example.
To do so, adjust mosaic signature and how to call its arguments with:
setMethod('mosaic', signature(x='list', y='missing'),
function(x, y, fun, tolerance=0.05, filename=""){
stopifnot(missing(y))
args <- x
if (!missing(fun)) args$fun <- fun
if (!missing(tolerance)) args$tolerance<- tolerance
if (!missing(filename)) args$filename<- filename
do.call(mosaic, args)
})
Let's keep tolerance low here to evaluate any misbehavior of our function.
Finally, the function:
Mosaic function
f.Mosaic <- function(x=x, func = median){
files <- list.files(file.path(wdata), all.files = F)
# List TIF files at wdata folder
ltif <- grep(".tif$", files, ignore.case = TRUE, value = TRUE)
#lext <- list()
#1rt <- raster(file.path(wdata, i),
# package = "raster", varname = fname, dataType = 'FLT4S')
# Give an extent area here (you can read it from your first tif or define manually)
uext <- extent(c(0, 100, 0, 100))
# Get Total Extent Area
stkl <- list()
for(i in 1:length(ltif)){
x <- raster(file.path(wdata, ltif[i]),
package = "raster", varname = fname, dataType = 'FLT4S')
xext <- extent(x)
uext <- union(uext, xext)
stkl[[i]] <- x
}
# Global Area empty rasterLayer
rt <- raster(uext)
res(rt) <- 0.5
rt[] <- NA
# Merge each rasterLayer to Global Extent area
stck <- list()
for(i in 1:length(stkl)){
merged.r <- merge(stkl[[i]], rt, tolerance = 1e+6)
#merged.r <- reclassify(merged.r, matrix(c(NA, 0), nrow = 1))
stck[[i]] <- merged.r
}
# Mosaic with Median
mosaic.r <- raster::mosaic(stck, fun = func) # using median
mosaic.r
}
# Run the function with func = median
mosaiced <- f.Mosaic(x, func = median)
# Plot it
plot(mosaiced)
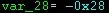
Possibly far from the best approach but hope it helps.