I would like to plot the result of fitted model on existing jpg. The result is between 0 and 1. And I would like the figure (as a pointer) to move nicely along the jpg axis (just about it as shown on the picture below).
I would like to plot the result of fitted model on existing jpg. The result is between 0 and 1. And I would like the figure (as a pointer) to move nicely along the jpg axis (just about it as shown on the picture below).

I'm not sure that I like the approach of using a ready made scale and adding a pointer to it; but, if you must, here is a way. The tricky part is getting your point plotting to line up with the axis displayed in your jpeg scale:
First, I made an example jpeg:
library(biOps)
#make jpeg of scale
jpeg("scale.jpeg", width=4, height=1, units="in", res=200, quality = 100)
par(mar=c(4,0,0,0), ps=10)
z <- seq(0,1,by=0.1)
breaks <- c(0,0.33,0.67,1)
image(x=z, z=matrix(z, nrow=length(z), ncol=1), xlim=c(0,1), axes=FALSE, xlab="", ylab="", breaks=breaks, col=c("red","yellow", "green"))
axis(1, at=seq(0.1,0.9,0.1))
box()
dev.off()
Then, I load this and place it in the lower panel of a device using layout, and added a point above at a given location (e.g. 0.2):
#add pointer above figure
pic <- readJpeg("scale.jpeg") # load jpeg
lo <- matrix(2:1, nrow=2, ncol=1) # layout info
WIDTHS <- c(4)
HEIGHTS <- c(0.2,1)
#make new jpeg
jpeg("scale_w_pointer.jpeg", width=4, height=1.2, units="in", res=200, quality = 100)
#quartz() # or x11() to preview
layout(lo, widths=WIDTHS, heights=HEIGHTS, respect=TRUE)
layout.show(2)
#plot 1
par(mar=c(0,0,0,0))
plot(pic)
#plot 2
par(mar=c(0,0,0,0))
plot(1, xlim=c(0,1), ylim=c(0,1), t="n", xaxs="i", yaxs="i", xlab="", ylab="", axes=FALSE)
points(0.2, 0.30, pch=25, bg=1, col=1)
dev.off()
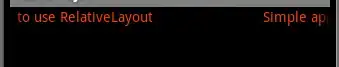
This is a bit rough but should get you started. With scan, tcltk or shiny you could make this interactive/responsive and/or write to file as needed.
col <- c("green", "yellow", "red")
x <- matrix(col, nrow = 1L)
plot(0, xlim = c(0, 1), ylim = c(0, 0.5), asp = 1,
xlab = "", ylab = "", axes = FALSE, type = "n")
axis(1, at = seq(0, 1, length = 6), line = -6, lwd = 0, lwd.tick = 1)
rasterImage(x, 0, 0, 1, 0.05, interpolate = FALSE)
p <- cbind(c(0, 0.015, -0.015, 0) + runif(1), c(0.05, 0.065, 0.065, 0.05))
polygon(p, col = "black")