I'm trying to calculate the clusters of a network using igraph in R, where all nodes are connected. The plot seems to work OK, but then I'm not able to return the correct groupings from my clusters.
In this example, the plot shows 4 main clusters, but in the largest cluster, not all nodes are connected:
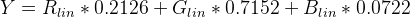
I would like to be able to return the following list of clusters from this graph object:
[[1]]
[1] 8 9
[[2]]
[1] 7 10
[[3]]
[1] 4 6 11
[[4]]
[1] 2 3 5
[[5]]
[1] 1 3 5 12
Example code:
library(igraph)
topology <- structure(list(N1 = c(1, 3, 5, 12, 2, 3, 5, 1, 2, 3, 5, 12, 4,
6, 11, 1, 2, 3, 5, 12, 4, 6, 11, 7, 10, 8, 9, 8, 9, 7, 10, 4,
6, 11, 1, 3, 5, 12), N2 = c(1, 1, 1, 1, 2, 2, 2, 3, 3, 3, 3,
3, 4, 4, 4, 5, 5, 5, 5, 5, 6, 6, 6, 7, 7, 8, 8, 9, 9, 10, 10,
11, 11, 11, 12, 12, 12, 12)), .Names = c("N1", "N2"), row.names = c(NA,
-38L), class = "data.frame")
g2 <- graph.data.frame(topology, directed=FALSE)
g3 <- simplify(g2)
plot(g3)
The cliques function gets me part of the way there:
tmp <- cliques(g3)
tmp
but, this list also gives groupings where not all nodes connect. For example, this clique includes the nodes 1,2,3,5 but 1 only connects to 3, and 2 only connects to 3 and 5, and 5 only connects to 2 :
topology[tmp[[31]],]
# N1 N2
#6 3 2
#7 5 2
#8 1 3
Thanks in advance for any help.