I am reading a dataset "Dummy_data.csv" with 26 columns and 4288 instances, where there are overall 17 parameters (columns) which are important for our data analysis. 6 out of 17 parameters i.e. (param1, param2, param3, param5, param6, param7) are critical parameters which being simultaneously out of range determines whether the item would be defective or not (class label). For example,
range1 = (min1, max1) = (0.25, 0.35)
range2 = (min2, max2) = (2.5, 3.1)
range3 = (min3, max3) = (680, 700)
range5 = (min5, max5) = (56, 64)
range6 = (min6, max6) = (40, 60)
range7 = (min7, max7) = (28, 38)
if (param1 out of range1 & param2 out of range2 & param3 out of range3 &
param5 out of range5 & param6 out of range6 & param7 out of range7)
class = 'defective'
else
class = 'ok'
We need to do two defect analyses on the above data. First, I need to find out the % share of defects from the total number of items. Second, I need to find out the frequency histogram of the out of range values for each of the 6 critical parameters to have an understanding of which out-of-range values of these critical parameters contributed more to defective items.
What I did:
Since the ranges of these 6 critical parameters were mostly non-overlapping, first I scaled the 17 parameters (though scaling the 6 critical parameters would have been sufficient!) using (x - min(x))/(max(x)-min(x))to an interval of (0, 1) so that I can do the frequency distribution of out of range values for 6 parameters on an uniform scale of x-axis. In graphical terms, this means a parameter value less than 0 means less than minimum value whereas a parameter value greater than 1 means more than maximum value. Thus, I filtered all the defective instances from the dataset in the z data frame and draw a pie chart to show percentage defective and OK items. (first analysis)
For the frequency histogram (second analysis), I filtered all the defective instances from the scaled dataset scaled.dat.df into defect.dat.df. Then I select the min and max from all the 6 parameters to determine the defect interval range. Next, I binned the unique values of each of the 6 parameters into p1.bin.defect.dat.df to p7.bin.defect.dat.df and plotted the individual histograms using the plot function on the same plot.
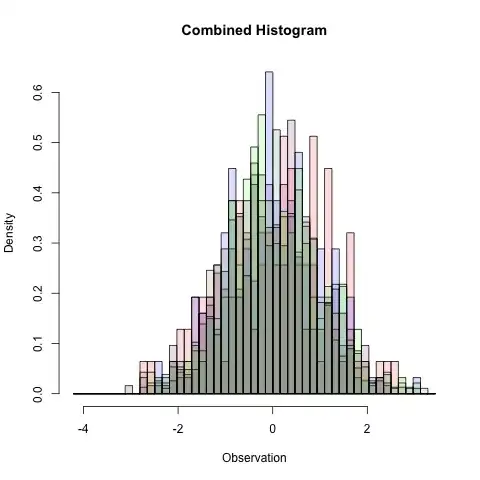
The problem with multiple overlapping histogram plots
I am getting the multi-histogram plot as shown below but the problem is that the width of the bars are varying for the 6 parameters. Does anybody have an idea of how to set a uniform bar width for a multi-histogram plot? Also, how can I add a suitable legend to this multi-hist plot ?
Any helpful suggestions/answers will be highly appreciated and rewarded accordingly.
Note: I followed the other thread on multiple histogram plot here how-to-plot-two-histograms-together-in-r and want a multi-hist plot very similar to this but 6 overlapping hist plots instead of 2 overlapping hist plots (as in the thread)
library(RWeka)
library(party)
library(plyr)
library(plotrix)
library(sm)
#read data and class labels
dat <- read.csv("Dummy_data.csv", head=T, sep=",")
datm <- as.matrix(dat[,8:24])
class <- as.matrix(dat[,26])
#center and scale data
center <- c(0.25, 2.5, 680, 1067, 56, 40, 28, -99, -99, 40, 5, 50, 5000, 15000, 11.3, 9.1, 0)
scale <- c(0.1, 0.6, 20, 6, 8, 20, 10, 19, 19, 20, 2, 10, 500, 1000, 3.4, 18.3, 5)
scaled.datm <- scale(datm, center, scale)
write.table(scaled.datm,
file = "C:\\Users\\schakrabarti\\Documents\\Dummy_data_whdr17.csv",
append=FALSE, quote=TRUE, sep=",", eol = "\n", na = "NA", dec = ".",
row.names = FALSE, col.names = TRUE, qmethod = c("escape", "double"),
fileEncoding = "")
#filter total non-compliants
scaled.dat.df <- as.data.frame(scaled.datm)
total <- length(scaled.dat.df[,1])
z <- c((scaled.dat.df[,"PARAM1"]<0 | scaled.dat.df[,"PARAM1"]>1) &
(scaled.dat.df[,"PARAM2"]<0 | scaled.dat.df[,"PARAM2"]>1) &
(scaled.dat.df[,"PARAM3"]<0 | scaled.dat.df[,"PARAM3"]>1) &
(scaled.dat.df[,"PARAM5"]<0 | scaled.dat.df[,"PARAM5"]>1) &
(scaled.dat.df[,"PARAM6"]<0 | scaled.dat.df[,"PARAM6"]>1) &
(scaled.dat.df[,"PARAM7"]<0 | scaled.dat.df[,"PARAM7"]>1) )
noncompliant <- length(z[z == TRUE])
slices <- c(noncompliant, total - noncompliant)
labls <- c("NOT OK","OK")
pct <- round(slices/sum(slices)*100, digits=2)
labls <- paste(labls, pct)
labls <- paste(labls, "%", sep="")
#pie3D(slices,labels=labls,explode=0.05, col=c(rgb(0.75,0,0.5),rgb(0,1,0.75)),main="Defect Analysis due to critical parameters")
pie(slices,labels=labls,main="Defect Analysis due to critical parameters")
#filter non-compliants due to individual params
defect.dat.df <- scaled.dat.df[z,]
#select defect interval range
min <- min(as.numeric(sapply(defect.dat.df[,c("PARAM1","PARAM2","PARAM3","PARAM5","PARAM6","PARAM7")], function(x) min(as.numeric(x)))))
max <- max(as.numeric(sapply(defect.dat.df[,c("PARAM1","PARAM2","PARAM3","PARAM5","PARAM6","PARAM7")], function(x) max(as.numeric(x)))))
#plot histogram for param1 defect
#p1.bin.defect.dat.df <- binning(defect.dat.df[,c("PARAM1")], breaks=seq(-0.4,0.2,by=0.2))
p1.bin.defect.dat.df <- binning(defect.dat.df[,c("PARAM1")], breaks=seq(min,max,by=0.2))
#h1 <- hist(defect.dat.df[,c("PARAM1")])
#plot(h1, col=rgb(1,0,0,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(head(p1.bin.defect.dat.df$breaks, n=1),tail(p1.bin.defect.dat.df$breaks, n=1)))
h1 <- hist(defect.dat.df[,c("PARAM1")], col=rgb(1,0,0,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(head(p1.bin.defect.dat.df$breaks, n=1),tail(p1.bin.defect.dat.df$breaks, n=1)))
#h1 <- hist(defect.dat.df[,c("PARAM1")], col=rgb(1,0,0,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(min,max))
box()
p2.bin.defect.dat.df <- binning(defect.dat.df[,c("PARAM2")], breaks=seq(min,max,by=0.2))
#h2 <- hist(defect.dat.df[,c("PARAM2")])
#plot(h2, col=rgb(0,0,1,1/7), xlim=c(head(p1.bin.defect.dat.df$breaks, n=1),tail(p1.bin.defect.dat.df$breaks, n=1)))
h2 <- hist(defect.dat.df[,c("PARAM2")], col=rgb(0,0,1,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(head(p2.bin.defect.dat.df$breaks, n=1),tail(p2.bin.defect.dat.df$breaks, n=1)), add=T)
#h2 <- hist(defect.dat.df[,c("PARAM2")], col=rgb(0,0,1,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(min,max), add=T)
box()
p3.bin.defect.dat.df <- binning(defect.dat.df[,c("PARAM3")], breaks=seq(min,max,by=0.2))
#h3 <- hist(defect.dat.df[,c("PARAM3")])
#plot(h3, col=rgb(0,1,0,1/7), xlim=c(head(p1.bin.defect.dat.df$breaks, n=1),tail(p1.bin.defect.dat.df$breaks, n=1)))
h3 <- hist(defect.dat.df[,c("PARAM3")], col=rgb(0,1,0,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(head(p3.bin.defect.dat.df$breaks, n=1),tail(p3.bin.defect.dat.df$breaks, n=1)), add=T)
#h3 <- hist(defect.dat.df[,c("PARAM3")], col=rgb(0,1,0,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(min,max), add=T)
box()
p5.bin.defect.dat.df <- binning(defect.dat.df[,c("PARAM5")], breaks=seq(min,max,by=0.2))
#h5 <- hist(defect.dat.df[,c("PARAM5")])
#plot(h5, col=rgb(0.5,0.5,0,1/7), xlim=c(head(p1.bin.defect.dat.df$breaks, n=1),tail(p1.bin.defect.dat.df$breaks, n=1)))
h5 <- hist(defect.dat.df[,c("PARAM5")], col=rgb(0.5,0,0.5,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(head(p5.bin.defect.dat.df$breaks, n=1),tail(p5.bin.defect.dat.df$breaks, n=1)), add=T)
#h5 <- hist(defect.dat.df[,c("PARAM5")], col=rgb(0.5,0,0.5,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(min,max), add=T)
box()
p6.bin.defect.dat.df <- binning(defect.dat.df[,c("PARAM6")], breaks=seq(min,max,by=0.2))
#h6 <- hist(defect.dat.df[,c("PARAM6")])
#plot(h6, col=rgb(0,0.5,0.5,1/7), xlim=c(head(p1.bin.defect.dat.df$breaks, n=1),tail(p1.bin.defect.dat.df$breaks, n=1)))
h6 <- hist(defect.dat.df[,c("PARAM6")], col=rgb(0,0.5,0.5,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(head(p6.bin.defect.dat.df$breaks, n=1),tail(p6.bin.defect.dat.df$breaks, n=1)), add=T)
#h6 <- hist(defect.dat.df[,c("PARAM6")], col=rgb(0,0.5,0.5,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(min,max), add=T)
box()
p7.bin.defect.dat.df <- binning(defect.dat.df[,c("PARAM7")], breaks=seq(min,max,by=0.2))
#h7 <- hist(defect.dat.df[,c("PARAM7")])
#plot(h7, col=rgb(0.5,0,0.5,1/7), xlim=c(head(p1.bin.defect.dat.df$breaks, n=1),tail(p1.bin.defect.dat.df$breaks, n=1)))
h7 <- hist(defect.dat.df[,c("PARAM7")], col=rgb(0.5,0.5,0,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(head(p7.bin.defect.dat.df$breaks, n=1),tail(p7.bin.defect.dat.df$breaks, n=1)), add=T)
#h7 <- hist(defect.dat.df[,c("PARAM7")], col=rgb(0.5,0.5,0,1/7), xlab="Param Defect Intervals", main="Frequency of Parameter Defects", xlim=c(min,max), add=T)
box()