the code shown here are simplied but triggers the same PicklingError. I know there is a lot discussion on what can and cannot be pickled, but I did find the solution from them.
I write a simple cython script with the following function:
def pow2(int a) :
return a**2
The compilation is working, I can call this function in python script.
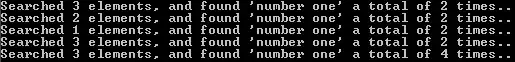
However, I am wondering how to use this function with multiprocessing,
from multiprocessing import Pool
from fast import pow2
p = Pool(processes =4 )
y = p.map( pow2, np.arange( 10, dtype=int))
gives me an PicklingError:
dtw is the name of the package, and fast is fast.pyx.
How can I get around this problem? Thanks in advance