I have a TOF spectrum and I would like to implement an algorithm using python (numpy) that finds all the maxima of the spectrum and returns the corresponding x values.
I have looked up online and I found the algorithm reported below.
The assumption here is that near the maximum the difference between the value before and the value at the maximum is bigger than a number DELTA. The problem is that my spectrum is composed of points equally distributed, even near the maximum, so that DELTA is never exceeded and the function peakdet returns an empty array.
Do you have any idea how to overcome this problem? I would really appreciate comments to understand better the code since I am quite new in python.
Thanks!
import sys
from numpy import NaN, Inf, arange, isscalar, asarray, array
def peakdet(v, delta, x = None):
maxtab = []
mintab = []
if x is None:
x = arange(len(v))
v = asarray(v)
if len(v) != len(x):
sys.exit('Input vectors v and x must have same length')
if not isscalar(delta):
sys.exit('Input argument delta must be a scalar')
if delta <= 0:
sys.exit('Input argument delta must be positive')
mn, mx = Inf, -Inf
mnpos, mxpos = NaN, NaN
lookformax = True
for i in arange(len(v)):
this = v[i]
if this > mx:
mx = this
mxpos = x[i]
if this < mn:
mn = this
mnpos = x[i]
if lookformax:
if this < mx-delta:
maxtab.append((mxpos, mx))
mn = this
mnpos = x[i]
lookformax = False
else:
if this > mn+delta:
mintab.append((mnpos, mn))
mx = this
mxpos = x[i]
lookformax = True
return array(maxtab), array(mintab)
Below is shown part of the spectrum. I actually have more peaks than those shown here.
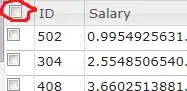
 For the noisy one, I filtered peaks with
For the noisy one, I filtered peaks with