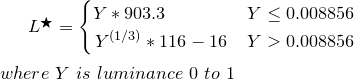I am trying matlab to plot ramachandran plot, without using built in command. I have succeeded too. Now I wanted to spot the GLYCINEs alone in the scatter array. Any ideas how to do this? (link to 1UBQ.pdb file : http://www.rcsb.org/pdb/download/downloadFile.do?fileFormat=pdb&compression=NO&structureId=1UBQ)
% Program to plot Ramanchandran plot of Ubiquitin
close all; clear ; clc; % close all figure windows, clear variables, clear screen
pdb1 ='/home/devanandt/Documents/VMD/1UBQ.pdb';
p=pdbread(pdb1); % read pdb file corresponding to ubiquitin protein
atom={p.Model.Atom.AtomName};
n_i=find(strcmp(atom,'N')); % Find indices of atoms
ca_i=find(strcmp(atom,'CA'));
c_i=find(strcmp(atom,'C'));
X = [p.Model.Atom.X];
Y = [p.Model.Atom.Y];
Z = [p.Model.Atom.Z];
X_n = X(n_i(2:end)); % X Y Z coordinates of atoms
Y_n = Y(n_i(2:end));
Z_n = Z(n_i(2:end));
X_ca = X(ca_i(2:end));
Y_ca = Y(ca_i(2:end));
Z_ca = Z(ca_i(2:end));
X_c = X(c_i(2:end));
Y_c = Y(c_i(2:end));
Z_c = Z(c_i(2:end));
X_c_ = X(c_i(1:end-1)); % the n-1 th C (C of cabonyl)
Y_c_ = Y(c_i(1:end-1));
Z_c_ = Z(c_i(1:end-1));
V_c_ = [X_c_' Y_c_' Z_c_'];
V_n = [X_n' Y_n' Z_n'];
V_ca = [X_ca' Y_ca' Z_ca'];
V_c = [X_c' Y_c' Z_c'];
V_ab = V_n - V_c_;
V_bc = V_ca - V_n;
V_cd = V_c - V_ca;
phi=0;
for k=1:numel(X_c)
n1=cross(V_ab(k,:),V_bc(k,:))/norm(cross(V_ab(k,:),V_bc(k,:)));
n2=cross(V_bc(k,:),V_cd(k,:))/norm(cross(V_bc(k,:),V_cd(k,:)));
x=dot(n1,n2);
m1=cross(n1,(V_bc(k,:)/norm(V_bc(k,:))));
y=dot(m1,n2);
phi=cat(2,phi,-atan2d(y,x));
end
phi=phi(1,2:end);
X_n_ = X(n_i(2:end)); % (n+1) nitrogens
Y_n_ = Y(n_i(2:end));
Z_n_ = Z(n_i(2:end));
X_ca = X(ca_i(1:end-1));
Y_ca = Y(ca_i(1:end-1));
Z_ca = Z(ca_i(1:end-1));
X_n = X(n_i(1:end-1));
Y_n = Y(n_i(1:end-1));
Z_n = Z(n_i(1:end-1));
X_c = X(c_i(1:end-1));
Y_c = Y(c_i(1:end-1));
Z_c = Z(c_i(1:end-1));
V_n_ = [X_n_' Y_n_' Z_n_'];
V_n = [X_n' Y_n' Z_n'];
V_ca = [X_ca' Y_ca' Z_ca'];
V_c = [X_c' Y_c' Z_c'];
V_ab = V_ca - V_n;
V_bc = V_c - V_ca;
V_cd = V_n_ - V_c;
psi=0;
for k=1:numel(X_c)
n1=cross(V_ab(k,:),V_bc(k,:))/norm(cross(V_ab(k,:),V_bc(k,:)));
n2=cross(V_bc(k,:),V_cd(k,:))/norm(cross(V_bc(k,:),V_cd(k,:)));
x=dot(n1,n2);
m1=cross(n1,(V_bc(k,:)/norm(V_bc(k,:))));
y=dot(m1,n2);
psi=cat(2,psi,-atan2d(y,x));
end
psi=psi(1,2:end);
scatter(phi,psi)
box on
axis([-180 180 -180 180])
title('Ramachandran Plot for Ubiquitn Protein','FontSize',16)
xlabel('\Phi^o','FontSize',20)
ylabel('\Psi^o','FontSize',20)
grid
The output is :
EDIT : Is my plot correct? Biopython: How to avoid particular amino acid sequences from a protein so as to plot Ramachandran plot? has an answer which has slightly different plot.
The modified code is as below :
% Program to plot Ramanchandran plot of Ubiquitin with no glycines
close all; clear ; clc; % close all figure windows, clear variables, clear screen
pdb1 ='/home/devanandt/Documents/VMD/1UBQ.pdb';
p=pdbread(pdb1); % read pdb file corresponding to ubiquitin protein
atom={p.Model.Atom.AtomName};
n_i=find(strcmp(atom,'N')); % Find indices of atoms
ca_i=find(strcmp(atom,'CA'));
c_i=find(strcmp(atom,'C'));
X = [p.Model.Atom.X];
Y = [p.Model.Atom.Y];
Z = [p.Model.Atom.Z];
X_n = X(n_i(2:end)); % X Y Z coordinates of atoms
Y_n = Y(n_i(2:end));
Z_n = Z(n_i(2:end));
X_ca = X(ca_i(2:end));
Y_ca = Y(ca_i(2:end));
Z_ca = Z(ca_i(2:end));
X_c = X(c_i(2:end));
Y_c = Y(c_i(2:end));
Z_c = Z(c_i(2:end));
X_c_ = X(c_i(1:end-1)); % the n-1 th C (C of cabonyl)
Y_c_ = Y(c_i(1:end-1));
Z_c_ = Z(c_i(1:end-1));
V_c_ = [X_c_' Y_c_' Z_c_'];
V_n = [X_n' Y_n' Z_n'];
V_ca = [X_ca' Y_ca' Z_ca'];
V_c = [X_c' Y_c' Z_c'];
V_ab = V_n - V_c_;
V_bc = V_ca - V_n;
V_cd = V_c - V_ca;
phi=0;
for k=1:numel(X_c)
n1=cross(V_ab(k,:),V_bc(k,:))/norm(cross(V_ab(k,:),V_bc(k,:)));
n2=cross(V_bc(k,:),V_cd(k,:))/norm(cross(V_bc(k,:),V_cd(k,:)));
x=dot(n1,n2);
m1=cross(n1,(V_bc(k,:)/norm(V_bc(k,:))));
y=dot(m1,n2);
phi=cat(2,phi,-atan2d(y,x));
end
phi=phi(1,2:end);
X_n_ = X(n_i(2:end)); % (n+1) nitrogens
Y_n_ = Y(n_i(2:end));
Z_n_ = Z(n_i(2:end));
X_ca = X(ca_i(1:end-1));
Y_ca = Y(ca_i(1:end-1));
Z_ca = Z(ca_i(1:end-1));
X_n = X(n_i(1:end-1));
Y_n = Y(n_i(1:end-1));
Z_n = Z(n_i(1:end-1));
X_c = X(c_i(1:end-1));
Y_c = Y(c_i(1:end-1));
Z_c = Z(c_i(1:end-1));
V_n_ = [X_n_' Y_n_' Z_n_'];
V_n = [X_n' Y_n' Z_n'];
V_ca = [X_ca' Y_ca' Z_ca'];
V_c = [X_c' Y_c' Z_c'];
V_ab = V_ca - V_n;
V_bc = V_c - V_ca;
V_cd = V_n_ - V_c;
psi=0;
for k=1:numel(X_c)
n1=cross(V_ab(k,:),V_bc(k,:))/norm(cross(V_ab(k,:),V_bc(k,:)));
n2=cross(V_bc(k,:),V_cd(k,:))/norm(cross(V_bc(k,:),V_cd(k,:)));
x=dot(n1,n2);
m1=cross(n1,(V_bc(k,:)/norm(V_bc(k,:))));
y=dot(m1,n2);
psi=cat(2,psi,-atan2d(y,x));
end
psi=psi(1,2:end);
res=strsplit(p.Sequence.ResidueNames,' ');
angle =[phi;psi];
angle(:,find(strcmp(res,'GLY'))-1)=[];
scatter(angle(1,:),angle(2,:))
box on
axis([-180 180 -180 180])
title('Ramachandran Plot for Ubiquitn Protein','FontSize',16)
xlabel('\Phi^o','FontSize',20)
ylabel('\Psi^o','FontSize',20)
grid
which gives output (with no GLY) as below :