I would like to ask you for the suggestions how I can edit my plot function to make my graph more clear ?
Here I show you the code which I use for plotting:
# open the pdf file
pdf(file='LSF1_PWD_GWD.pdf')
a <- c('LSF1', 'PWD', 'GWD')
rowsToPlot<-c(1066,2269,109)
matplot(as.matrix(t(tbl_alles[rowsToPlot,])),type=rep("l", length(rowsToPlot)), col=rainbow(length(rowsToPlot)),xlab = 'Fraction Size', ylab = 'Intensity')
legend('topright',a,lty=1, bty='n', cex=.75, col = rainbow(length(rowsToPlot)))
# close the pdf file
dev.off()
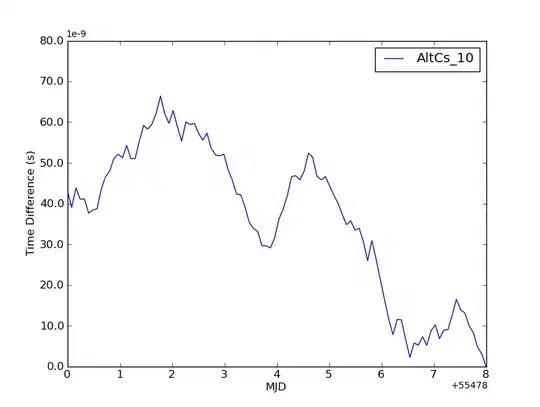
and that's how the graph looks like:
It's just a basic plot because I have no idea how to edit it. The arrow indicates three lines on one position which you can't see because they overlap... and that's the most important part of this graph for me. Maybe I shouldn't use dotted line ? How to change it ?
Data:
tbl_alles <-
structure(list("10" = c(0, 0, 0, 0, 0, 0),
"20" = c(0, 0, 0, 0, 0, 0),
"52.5" = c(0, 0, 0, 0, 0, 0),
"81" = c(0, 0, 1, 0, 0, 0),
"110" = c(0, 0, 0, 0, 0, 0),
"140.5" = c(0, 0, 0, 0, 0, 0),
"189" = c(0, 0, 0, 0, 0, 0),
"222.5" = c(0, 0, 0, 0, 0, 0 ),
"278" = c(0, 0, 0, 0, 0, 0),
"340" = c(0, 0, 0, 0, 0, 0),
"397" = c(0, 1, 0, 0, 0, 0),
"453.5" = c(0, 0.66069369, 0, 0, 0, 1),
"529" = c(0, 0.521435654, 0, 0, 1, 0),
"580" = c(0, 0.437291195, 0, 0, 1, 0),
"630.5" = c(0, 0.52204783, 0, 0, 0, 0),
"683.5" = c(0, 0.52429838, 0, 0, 0, 0),
"735.5" = c(1, 0.3768651, 0, 1, 0, 0),
"784" = c(0, 0, 0, 0, 0, 0),
"832" = c(0, 0, 0, 0, 0, 0),
"882.5" = c(0, 0, 0, 0, 0, 0),
"926.5" = c(0, 0, 0, 0, 0, 0),
"973" = c(0, 0, 0, 0, 0, 0),
"1108" = c(0, 0, 0, 0, 0, 0),
"1200" = c(0, 0, 0, 0, 0, 0)),
.Names = c("10", "20", "52.5", "81",
"110", "140.5","189", "222.5",
"278", "340", "397", "453.5",
"529", "580", "630.5", "683.5",
"735.5", "784", "832", "882.5",
"926.5", "973", "1108", "1200"),
row.names = c("at1g01050.1", "at1g01080.1",
"at1g01090.1","at1g01220.1",
"at1g01420.1", "at1g01470.1"),
class = "data.frame")
RowsToPlot:
> dput(tbl_alles[rowsToPlot,])
structure(list(`10` = c(0, 0, 0), `20` = c(0, 0, 0), `52.5` = c(0,
0, 0), `81` = c(0, 0, 0), `110` = c(0, 0, 0), `140.5` = c(0,
0, 0), `189` = c(0, 0, 0), `222.5` = c(0, 0, 0), `278` = c(0,
0, 0), `340` = c(0, 0, 0), `397` = c(0, 0, 0), `453.5` = c(0,
0, 0), `529` = c(0, 0, 0), `580` = c(0, 0, 0), `630.5` = c(0,
0, 0), `683.5` = c(0, 0, 0.57073483), `735.5` = c(0, 1, 0.85691826
), `784` = c(0, 0, 0.90706982), `832` = c(1, 1, 1), `882.5` = c(0,
0, 0), `926.5` = c(0, 0, 0), `973` = c(0, 0, 0), `1108` = c(0,
0, 0), `1200` = c(0, 0, 0)), .Names = c("10", "20", "52.5", "81",
"110", "140.5", "189", "222.5", "278", "340", "397", "453.5",
"529", "580", "630.5", "683.5", "735.5", "784", "832", "882.5",
"926.5", "973", "1108", "1200"), row.names = c("at3g01510.1",
"at5g26570.1", "at1g10760.1"), class = "data.frame")