I see several people have answered the question for plotting with an irregular grid. I'm having trouble getting the contour lines to line up with the filled contours. Also, need to display the data point locations on the plot, and the radial spokes at 30 deg increments, and semi circles at 10, 20 30.
Ref: Plotting contours on an irregular grid
heading=seq(0,180,30)
speed=c(5,10,15,20,30)
mheading=matrix(heading,ncol=length(heading),nrow=length(speed),byrow=TRUE)
mspeed=matrix(speed,ncol=length(heading),nrow=length(speed),byrow=FALSE)
mag=mheading+mspeed
x=sin(mheading*pi/180)*mspeed
y=cos(mheading*pi/180)*mspeed
z=mag
library(akima)
df<-data.frame(x=x,y=y,z=z)
# interpolation
fld <- with(df, interp(x = x, y = y, z = z,
xo=seq(min(x),max(x),length=100),
yo=seq(min(y),max(y),length=100)))
filled.contour(x = fld$x,
y = fld$y,
z = fld$z,
color.palette =
colorRampPalette(c("white", "blue")),
xlab = "",
ylab = "",
main = "Max",
key.title = title(main = "Value", cex.main = 1),
asp=1,xlim=c(0,40),ylim=c(-30,30))
contour(x = fld$x,
y = fld$y,
z = fld$z,
color.palette =
colorRampPalette(c("white", "blue")),
xlab = "",
ylab = "",
asp=1,xlim=c(0,40),ylim=c(-30,30), add=TRUE)
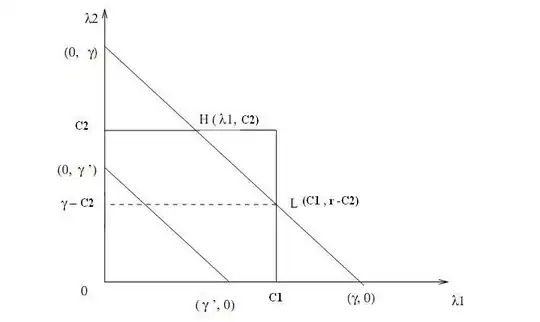
Following this link, produces the following code / plot. This is "better", but there are still problems. Why is there interpolated data inside of the minimum speed radius (5), and why doesn't the contour fill/lines extend to the outer radius especially near 90 degrees?
contours=TRUE # Add contours to the plotted surface
legend=TRUE # Plot a surface data legend?
axes=TRUE # Plot axes?
points=TRUE # Plot individual data points
extrapolate=FALSE # Should we extrapolate outside data points?
single_point_overlay=0
outer.radius=30
spatial_res=1000 #Resolution of fitted surface
interp.type = 1
circle.rads <- pretty(c(0,outer.radius))
heading=seq(0,180,30)
speed=c(5,10,15,20,30)
mheading=matrix(heading,ncol=length(heading),nrow=length(speed),byrow=TRUE)
mspeed=matrix(speed,ncol=length(heading),nrow=length(speed),byrow=FALSE)
mag=mheading+mspeed
x=sin(mheading*pi/180)*mspeed
y=cos(mheading*pi/180)*mspeed
z=mag
extrapolate=FALSE # Should we extrapolate outside data points?
contour_levels = 8
col_levels=contour_levels
col_breaks_source=1
contour_breaks_source = 1
col = rev(heat.colors(col_levels))
minitics <- seq(-outer.radius, outer.radius, length.out = spatial_res)
xmini <- seq(min(x),max(x),length=spatial_res)
ymini <- seq(min(y),max(y),length=spatial_res)
# interpolate the data
if (interp.type ==1 ){
# Interp <- akima:::interp(x = x, y = y, z = z,
# extrap = extrapolate,
# xo = xmini,
# yo = ymini,
# linear = FALSE)
# Mat <- Interp[[3]]
df<-data.frame(x=x,y=y,z=z)
# interpolation
fld <- with(df, akima:::interp(x = x, y = y, z = z,
xo=xmini,
yo=ymini))
Mat_x <- fld[[1]]
Mat_y <- fld[[2]]
Mat_z <- fld[[3]]
} else if (interp.type == 2){
library(fields)
grid.list = list(x=minitics,y=minitics)
t = Tps(cbind(x,y),z,lambda=lambda)
tmp = predict.surface(t,grid.list,extrap=extrapolate)
Mat_z = tmp$z
# mark cells outside circle as NA
markNA <- matrix(minitics, ncol = spatial_res, nrow = spatial_res)
Mat_x <- markNA
Mat_y <- t(markNA)
} else {stop("interp.type value not valid")}
#
Mat_z[!(sqrt(Mat_x ^ 2 + Mat_y ^ 2) <= max(speed)*1.1)] <- NA
Mat_z[!(sqrt(Mat_x ^ 2 + Mat_y ^ 2) >= min(speed))] <- NA # <- SHOULD REMOVE INNER DATA
### Set contour_breaks based on requested source
if ((length(contour_breaks_source == 1)) & (contour_breaks_source[1] == 1)){
# contour_breaks = seq(min(z,na.rm=TRUE),max(z,na.rm=TRUE),
# by=(max(z,na.rm=TRUE)-min(z,na.rm=TRUE))/(contour_levels-1))
contour_breaks = seq(min(z,na.rm=TRUE),max(z,na.rm=TRUE),length.out = contour_levels+1)
}else if ((length(contour_breaks_source == 1)) & (contour_breaks_source[1] == 2)){
contour_breaks = seq(min(Mat_z,na.rm=TRUE),max(Mat_z,na.rm=TRUE),
by=(max(Mat_z,na.rm=TRUE)-min(Mat_z,na.rm=TRUE))/(contour_levels-1))
} else if ((length(contour_breaks_source) == 2) & (is.numeric(contour_breaks_source))){
contour_breaks = pretty(contour_breaks_source,n=contour_levels)
contour_breaks = seq(contour_breaks_source[1],contour_breaks_source[2],
by=(contour_breaks_source[2]-contour_breaks_source[1])/(contour_levels-1))
}else {stop("Invalid selection for \"contour_breaks_source\"")}
### Set color breaks based on requested source
if ((length(col_breaks_source) == 1) & (col_breaks_source[1] == 1))
{zlim=c(min(z,na.rm=TRUE),max(z,na.rm=TRUE))} else if ((length(col_breaks_source) == 1) & (col_breaks_source[1] == 2))
{zlim=c(min(Mat_z,na.rm=TRUE),max(Mat_z,na.rm=TRUE))} else if ((length(col_breaks_source) == 2) & (is.numeric(col_breaks_source)))
{zlim=col_breaks_source} else {stop("Invalid selection for \"col_breaks_source\"")}
# begin plot
Mat_plot = Mat_z
Mat_plot[which(Mat_plot<zlim[1])]=zlim[1]
Mat_plot[which(Mat_plot>zlim[2])]=zlim[2]
image(x = Mat_x, y = Mat_y, Mat_plot ,
useRaster = TRUE, asp = 1, axes = FALSE, xlab = "", ylab = "", zlim = zlim, col = col)
# add contours if desired
if (contours){
CL <- contourLines(x = Mat_x, y = Mat_y, Mat_z, levels = contour_breaks)
A <- lapply(CL, function(xy){
lines(xy$x, xy$y, col = gray(.2), lwd = .5, asp=1)
})
}
# add interpolated point if desired
if (points){
points(x,y,pch=4)
}
# add overlay point (used for trained image marking) if desired
if (single_point_overlay!=0){
points(x[single_point_overlay],y[single_point_overlay],pch=0)
}
# add radial axes if desired
if (axes){
# internals for axis markup
RMat <- function(radians){
matrix(c(cos(radians), sin(radians), -sin(radians), cos(radians)), ncol = 2)
# matrix(c(sin(radians), -cos(radians), cos(radians), sin(radians)), ncol = 2)
}
circle <- function(x, y, rad = 1, nvert = 500, angle=360){
rads <- seq(0,angle*pi/180,length.out = nvert)
# xcoords <- cos(rads) * rad + x
# ycoords <- sin(rads) * rad + y
xcoords=sin(rads)*rad + x
ycoords=cos(rads)*rad + y
cbind(xcoords, ycoords)
}
# draw circles
if (missing(circle.rads)){
circle.rads <- pretty(c(0,outer.radius))
}
endAngle = 180
for (i in circle.rads){
lines(circle(0, 0, i, angle = endAngle), col = "#66666650")
}
# put on radial spoke axes:
axis.degs <- c(0, 30, 60, 90, 120, 150)
# axis.rads <- c(0, pi / 6, pi / 3, pi / 2, 2 * pi / 3, 5 * pi / 6)
axis.rads <- axis.degs * pi/180
r.labs <- c(90, 60, 30, 0, 330, 300)
l.labs <- c(270, 240, 210, 180, 150, 120)
for (i in 1:length(axis.rads)){
if (axis.degs[i]==0) {
# endpoints <- zapsmall(c(RMat(axis.rads[i]) %*% matrix(c(1, 0, -1, 0) * outer.radius,ncol = 2)))
endpoints <- zapsmall(c(RMat(axis.rads[i]) %*% matrix(c(1, 0, 0, 0) * outer.radius,ncol = 2)))
} else if (0 < axis.degs[i] & axis.degs[i] < 90) {
endpoints <- zapsmall(c(RMat(axis.rads[i]) %*% matrix(c(1, 0, 0, 0) * outer.radius,ncol = 2)))
} else {
endpoints <- zapsmall(c(RMat(axis.rads[i]) %*% matrix(c(0, 0, -1, 0) * outer.radius,ncol = 2)))
}
segments(endpoints[1], endpoints[2], endpoints[3], endpoints[4], col = "#66666650")
endpoints <- c(RMat(axis.rads[i]) %*% matrix(c(1.1, 0, -1.1, 0) * outer.radius, ncol = 2))
lab1 <- bquote(.(r.labs[i]) * degree)
lab2 <- bquote(.(l.labs[i]) * degree)
if (0 <= r.labs[i] & r.labs[i] <= 180) text(endpoints[1], endpoints[2], lab1, xpd = TRUE)
if (0 <= l.labs[i] & l.labs[i] <= 180) text(endpoints[3], endpoints[4], lab2, xpd = TRUE)
}
# axis(2, pos = -1.25 * outer.radius, at = sort(union(circle.rads,-circle.rads)), labels = NA)
# text( -1.26 * outer.radius, sort(union(circle.rads, -circle.rads)),sort(union(circle.rads, -circle.rads)), xpd = TRUE, pos = 2)
axis(2, pos = 0 * outer.radius, at = sort(union(circle.rads,-circle.rads)), labels = NA)
text( -0.02 * outer.radius, sort(union(circle.rads, -circle.rads)),
abs(sort(union(circle.rads, -circle.rads))),
xpd = TRUE, pos = 2)
}
# add legend if desired
# this could be sloppy if there are lots of breaks, and that's why it's optional.
# another option would be to use fields:::image.plot(), using only the legend.
# There's an example for how to do so in its documentation
if (legend){
library(fields)
image.plot(legend.only=TRUE, smallplot=c(.78,.82,.1,.8), col=col, zlim=zlim)
# ylevs <- seq(-outer.radius, outer.radius, length = contour_levels+ 1)
# #ylevs <- seq(-outer.radius, outer.radius, length = length(contour_breaks))
# rect(1.2 * outer.radius, ylevs[1:(length(ylevs) - 1)], 1.3 * outer.radius, ylevs[2:length(ylevs)], col = col, border = NA, xpd = TRUE)
# rect(1.2 * outer.radius, min(ylevs), 1.3 * outer.radius, max(ylevs), border = "#66666650", xpd = TRUE)
# text(1.3 * outer.radius, ylevs[seq(1,length(ylevs),length.out=length(contour_breaks))],round(contour_breaks, 1), pos = 4, xpd = TRUE)
}