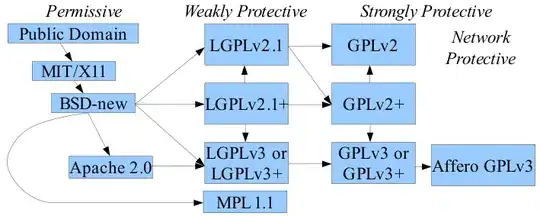
Use the smooth.spline function (Fit a Smoothing Spline)
mx=path.w.missing$x
my=path.w.missing$y
s<-smooth.spline(mx,my)
plot(mx,my)
lines(s) #draw the estimated path
predict(s,1:10) #And now you can generate all the points on the estimated path
> predict(s,1:10)
$x
[1] 1 2 3 4 5 6 7 8 9 10
$y
[1] 2.418294 2.904019 3.389744 3.875469 4.361195 4.846920 5.332645 5.818370 6.304095 6.789820
And you can use the Linear model to fit it.
> summary(lm(my~mx))
Call:
lm(formula = my ~ mx)
Residuals:
Min 1Q Median 3Q Max
-12.772 -8.642 3.112 5.831 17.237
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -12.60098 3.46583 -3.636 0.000444 ***
mx 0.56579 0.02138 26.469 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 7.896 on 98 degrees of freedom
Multiple R-squared: 0.8773, Adjusted R-squared: 0.876
F-statistic: 700.6 on 1 and 98 DF, p-value: < 2.2e-16
as you can see,p<0.05,so my and mx can fit as a line:
my=-12.60098+0.56579*mx
ggplot2 can draw this line easily:
d=data.frame(mx,my)
library(ggplot2)
ggplot(d,aes(mx,my))+geom_point()+geom_smooth(method="lm")

assume that a line is Ax+By+C=0 and the point is(X0,Y0),point to the line distance:
|AX0+BY0+C|/√(A²+B²)
so in this case, 0.56579*mx-my-12.60098=0 ,A=0.56579,B=-1,C=-12.60098 ,it's easy to calculate the distance from point to line and find the closest point to the line.
Further more,if you want to find the closest point , remove the denominator √(A²+B²) would not affect the sort,so,the optimization of the formula:
|AX0+BY0+C|
Result
> for(i in 1:2503501){
+ temp=abs(centers[[1]][i]*0.56579-centers[[2]][i]-12.60098)
+ if(m>temp){
+ m=temp
+ pos=i
+ }
+ }
> m
[1] 2.523392e-05
> pos
[1] 638133
Use the Rcpp to accelerate the program "test.cpp"
#include <Rcpp.h>
using namespace Rcpp;
// [[Rcpp::export]]
int closest(NumericVector a, NumericVector b) {
int length = a.size();
double temp,m=10000;
int pos=0;
for(int i=0;i<length;i++){
temp=a[i]*0.56579-b[i]-12.60098;
if(temp>=0){
if(m>temp){m=temp;pos=i;}
}else{
if(m>-temp){m=-temp;pos=i;}
}
}
return pos+1;
}
Result(the Rcpp cost about 2 seconds):
> sourceCpp("test.cpp")
> closest(centers[[1]],centers[[2]])
[1] 974698
> abs(centers[[1]][974698]*0.56579-centers[[2]][974698]-12.60098)
[1] 0.0002597022