I am having some serious problems with layout() and it is driving me crazy when adding one figure with multiple layers.
I only seem to have a problem when adding a layer on one of the figures within the jpeg I am trying to create.
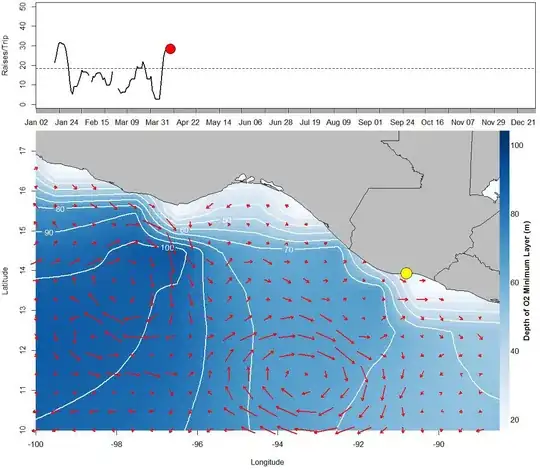
The layout is to have a 1) simple line plot on top for a time series of fish catch data and on the bottom 2) have a larger image of a map of oceanographic data layers.
I am making a series of maps using image.plot() plus contour(... add=T) and arrows my.symbols(... add=T) from library TeachingDemos.
Data is sliced from large netCDF files.
The image and contours are dissolved oxygen depth and the red arrows are surface current.
Below is my R code, data is looped through variable 'n':
jpeg(paste(interpdate[n],"DailyDOLayerCenAm.jpg", sep=""), width=1150, height=1000, res=100)
layout(matrix(c(1,2),nrow=2), heights=c(1,3))
#first plot on the top, fish catch data by time, moving each day
par(mar=c(1,4,.3,.5))
plot(Date[15:n],sail$X7.day.Average[15:n], xlim=c(Date[15],Date[350]),
xlab='',ylab='Raises/Trip',ylim=c(0,50), type='l', xaxt='n', lwd=2.5)
axis(1, Date, format(Date, "%b %d"), cex.axis = 1)
abline(18.4,0, lty=2)
points(Date[n],sail$X7.day.Average[n], pch=21, col='black', bg='red',
cex=3)
#second plot, Ocean data
par(mar=c(3,3.7,.5,1))
# layer 1 plot the main layer, interpolated grid O2 minimum depth
image.plot( as.surface( expandgrid, ww),xlim=c(xmin,xmax),
ylim=c(ymin,ymax), ylab="Latitude", xlab="Longitude",main="",
col=pal(256), legend.lab="Depth of O2 Minimum Layer (m)",
zlim=c(20,zlimit), cex=1.5)
#layer 2 add the contours
contour(as.surface( expandgrid, ww),xlim=c(xmin,xmax), ylim=c(ymin,ymax),
col='white', lwd=2, nlevels=10, labcex=1, add=T)
#layer 3 add current arrows
my.symbols(lonx,laty,ms.arrows, angle=theta, r=intensity, length=.06,
add=T,xlim=c(xmin,xmax), ylim=c(ymin,ymax), lwd=2, col="red",
fg="black")
#layer 4add the map of land/countries
plot(newmap, col="GREY", add=T)
#add a point of home port in Guatemala
points(-90.81, 13.93, pch=21, col='black', bg='yellow', cex=3.5)
dev.off()
When I plot just my ocean data, it plots fine: https://fbcdn-sphotos-f-a.akamaihd.net/hphotos-ak-xap1/t31.0-8/10861076_10101738319375937_3436888929444896713_o.jpg
plot it within layout() I get this mess, The contour lines are fine within the x-space, but squished in y space, as are the arrows and map overlay: https://scontent-b.xx.fbcdn.net/hphotos-xfp1/t31.0-8/10923795_10101738319625437_7583695382864373718_o.jpg