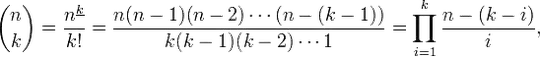I have following function:
def get_denom(n_comp,qs,x,cp,cs):
'''
len(n_comp) = 1 # number of proteins
len(cp) = n_comp # protein concentration
len(qp) = n_comp # protein capacity
len(x) = 3*n_comp + 1 # fit parameters
len(cs) = 1
'''
k = x[0:n_comp]
sigma = x[n_comp:2*n_comp]
z = x[2*n_comp:3*n_comp]
a = (sigma + z)*( k*(qs/cs)**(z-1) )*cp
denom = np.sum(a) + cs
return denom
I compare it against a Fortran implementation (My first Fortran function ever):
subroutine get_denom (qs,x,cp,cs,n_comp,denom)
! Calculates the denominator in the SMA model (Brooks and Cramer 1992)
! The function is called at a specific salt concentration and isotherm point
! I loops over the number of components
implicit none
! declaration of input variables
integer, intent(in) :: n_comp ! number of components
double precision, intent(in) :: cs,qs ! salt concentration, free ligand concentration
double precision, dimension(n_comp), INTENT(IN) ::cp ! protein concentration
double precision, dimension(3*n_comp + 1), INTENT(IN) :: x ! parameters
! declaration of local variables
double precision, dimension(n_comp) :: k,sigma,z
double precision :: a
integer :: i
! declaration of outpur variables
double precision, intent(out) :: denom
k = x(1:n_comp) ! equlibrium constant
sigma = x(n_comp+1:2*n_comp) ! steric hindrance factor
z = x(2*n_comp+1:3*n_comp) ! charge of protein
a = 0.
do i = 1,n_comp
a = a + (sigma(i) + z(i))*(k(i)*(qs/cs)**(z(i)-1.))*cp(i)
end do
denom = a + cs
end subroutine get_denom
I compiled the .f95 file by using:
1) f2py -c -m get_denom get_denom.f95 --fcompiler=gfortran
2) f2py -c -m get_denom_vec get_denom.f95 --fcompiler=gfortran --f90flags='-msse2' (The last option should turn on auto-vectorization)
I test the functions by:
import numpy as np
import get_denom as fort_denom
import get_denom_vec as fort_denom_vec
from matplotlib import pyplot as plt
%matplotlib inline
def get_denom(n_comp,qs,x,cp,cs):
k = x[0:n_comp]
sigma = x[n_comp:2*n_comp]
z = x[2*n_comp:3*n_comp]
# calculates the denominator in Equ 14a - 14c (Brooks & Cramer 1992)
a = (sigma + z)*( k*(qs/cs)**(z-1) )*cp
denom = np.sum(a) + cs
return denom
n_comp = 100
cp = np.tile(1.243,n_comp)
cs = 100.
qs = np.tile(1100.,n_comp)
x= np.random.rand(3*n_comp+1)
denom = np.empty(1)
%timeit get_denom(n_comp,qs,x,cp,cs)
%timeit fort_denom.get_denom(qs,x,cp,cs,n_comp)
%timeit fort_denom_vec.get_denom(qs,x,cp,cs,n_comp)
I added following Cython code:
import cython
# import both numpy and the Cython declarations for numpy
import numpy as np
cimport numpy as np
@cython.boundscheck(False)
@cython.wraparound(False)
def get_denom(int n_comp,np.ndarray[double, ndim=1, mode='c'] qs, np.ndarray[double, ndim=1, mode='c'] x,np.ndarray[double, ndim=1, mode='c'] cp, double cs):
cdef int i
cdef double a
cdef double denom
cdef double[:] k = x[0:n_comp]
cdef double[:] sigma = x[n_comp:2*n_comp]
cdef double[:] z = x[2*n_comp:3*n_comp]
# calculates the denominator in Equ 14a - 14c (Brooks & Cramer 1992)
a = 0.
for i in range(n_comp):
#a += (sigma[i] + z[i])*( pow( k[i]*(qs[i]/cs), (z[i]-1) ) )*cp[i]
a += (sigma[i] + z[i])*( k[i]*(qs[i]/cs)**(z[i]-1) )*cp[i]
denom = a + cs
return denom
EDIT:
Added Numexpr, using one thread:
def get_denom_numexp(n_comp,qs,x,cp,cs):
k = x[0:n_comp]
sigma = x[n_comp:2*n_comp]
z = x[2*n_comp:3*n_comp]
# calculates the denominator in Equ 14a - 14c (Brooks & Cramer 1992)
a = ne.evaluate('(sigma + z)*( k*(qs/cs)**(z-1) )*cp' )
return cs + np.sum(a)
ne.set_num_threads(1) # using just 1 thread
%timeit get_denom_numexp(n_comp,qs,x,cp,cs)
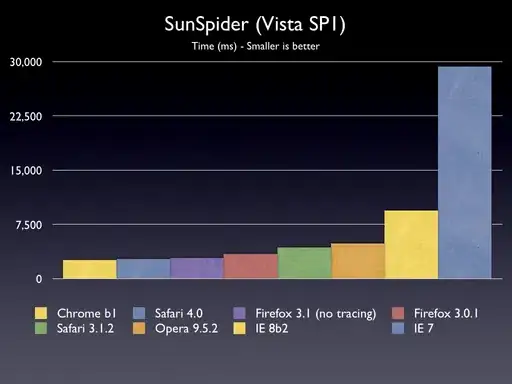
The result is (smaller is better):
Why is is the speed of Fortran getting closer to Numpy with increasing size of the arrays? And how could i speed up Cython? Using pointers?