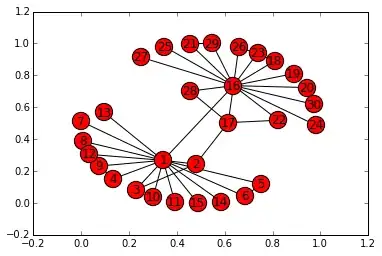
I have a graph network data (using adjacency matrix) of 30 nodes. The graph currently looks like this:
Each cluster has 15 nodes and each node is connected to other node within the same cluster. Only two pair of nodes of different cluster connect with each other. The problem is the graph I am getting is all condensed and each edges within the cluster is not clearly visible. Is there a way I can clearly show each edge within the cluster. Mainly like make the graph bigger with clear edge lines of each node visible.
I plotted this using the following command of networkx lib.
G1=nx.from_numpy_matrix(W1)
nx.draw_networkx(G1)
Where W1 is the adjacency matrix (30x30) of the nodes.
Please advise.
EDIT:
Want something like this with each node clear and edge visible and not condensed. The point is I want the upper cluster points to appear as near that cluster only and same for lower cluster points. But within each cluster, I want the nodes to be a bit separate so that each edge is visible clearly.

EDIT2:
def adjacencyMatrix2():
for x in range(N):
if (x<15):
c=N/2
else:
c=N
for y in range(x+1,c):
W1[x][y]=W1[y][x]=1
# Connecting two other nodes separately.
W1[0][16]=W1[16][0]=1
W1[1][15]=W1[15][1]=1
adjacencyMatrix2()
G1=nx.from_numpy_matrix(W1)
graph_pos=nx.spring_layout(G1,k=0.50,iterations=50)
nx.draw_networkx(G1,graph_pos)
EDIT3:
N=30
# Creating a matrix of zeros.
W=np.zeros((N,N))
# Mentioning the edges to start with. Thinking of a pair of 15 node cluster with two cluster connected by two pair of nodes.
edge=[[1,2],[1,3],[1,4],[1,5],[1,6],[1,7],[1,8],[1,9],[1,10],[1,11],[1,12],[1,13],[1,14],[1,15],
[16,17],[16,18],[16,19],[16,20],[16,21],[16,22],[16,23],[16,24],[16,25],[16,26],[16,27],[16,28],[16,29],[16,30],
[1,16],[2,17],[2,3],[5,6],[8,9],[9,4],[18,26],[17,22],[29,21],[17,28]]
# Function for creating adjacency matrix ,populating the zeros matrix with 1 and 0-signifying edges on a node.
def adjacencyMatrix():
"""This function creates an Adjacency Matrix from a edge set.
input-> set of edges to be connected
output-> Adjacency matrix (n,n)
"""
for first,second in edge:
W[first-1,second-1]=W[second-1][first-1]=1
# Creating the adjacency matrix by calling the function.
adjacencyMatrix()
Also I have seen for each run of the code the layout of the graph changes. I don't want that the graph layout to change with each run of the code. Currently it is doing that.