Okay, I found out the real problem: when you create the histogram with those bin-edge settings, the histogram creates bars which have equal size, and equal outside-spacing on the non-log scale.
To demonstrate, here's a zoomed-in version of the plot in the question, but in non-log scale:
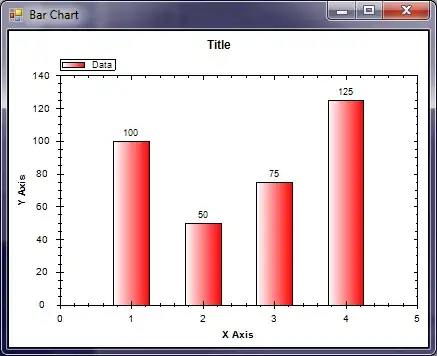
Notice how the first two bars are centered around (0 + 0.1) / 2 = 0.05, with a gap of 0.1 / 10 = 0.01 at the edges, while the next two bars are centered around (0.1 + 1.0) / 2 = 0.55, with a gap of 1.1 / 10 = 0.11 at either edge.
When converting things to log scale, bar widths and edge widths all go for a huge toss. This is compounded further by the fact that you have a linear scale from 0 to 0.1, after which things become log-scale.
I know no way of fixing this, other than to do everything manually. I've used the geometric means of the bin-edges in order to compute what the bar edges and bar widths should be. Note that this piece of code will work only for two datasets. If you have more datasets, you'll need to have some function that fills in the bin-edges with a geometric series appropriately.
import numpy as np
import matplotlib.pyplot as plt
def geometric_means(a):
"""Return pairwise geometric means of adjacent elements."""
return np.sqrt(a[1:] * a[:-1])
a = [0.05, 0.1, 0.2, 1, 2, 3] * 2
b = [0.05, 0.1, 0.2, 1, 2, 3] * 3
# Find frequencies
bins = np.array([0, 0.1, 1, 10])
a_hist = np.histogram(a, bins=bins)[0]
b_hist = np.histogram(b, bins=bins)[0]
# Find log-scale mid-points for bar-edges
mid_vals = np.hstack((np.array([0.05,]), geometric_means(bins[1:])))
# Compute bar left-edges, and bar widths
a_x = np.empty(mid_vals.size * 2)
a_x = bins[:-1]
a_widths = mid_vals - bins[:-1]
b_x = np.empty(mid_vals.size * 2)
b_x = mid_vals
b_widths = bins[1:] - mid_vals
plt.bar(a_x, a_hist, width=a_widths, color='b')
plt.bar(b_x, b_hist, width=b_widths, color='g')
plt.gca().set_xscale("symlog", linthreshx=0.1)
plt.show()
And the final result:

Sorry, but the neat gaps between the bars get killed. Again, this can be fixed by doing the appropriate geometric interpolation, so that everything is linear on log-scale.