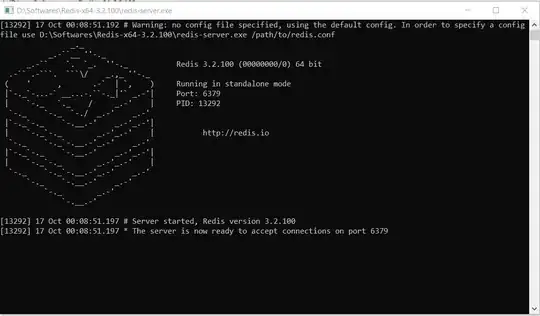
With version 1.37.1 (released in May 2020), texreg introduces the threeparttable argument, which uses the threeparttable LaTeX package, which was designed for this purpose.
Example R code:
texreg(lm(speed ~ dist, data = cars),
custom.note = paste("\\item %stars. This regression",
"should be interpreted with strong",
"caution as it is likely plagued by",
"extensive omitted variable bias."),
single.row = TRUE,
threeparttable = TRUE)
Output:
\begin{table}
\begin{center}
\begin{threeparttable}
\begin{tabular}{l c}
\hline
& Model 1 \\
\hline
(Intercept) & $8.28 \; (0.87)^{***}$ \\
dist & $0.17 \; (0.02)^{***}$ \\
\hline
R$^2$ & $0.65$ \\
Adj. R$^2$ & $0.64$ \\
Num. obs. & $50$ \\
\hline
\end{tabular}
\begin{tablenotes}[flushleft]
\scriptsize{\item $^{***}p<0.001$; $^{**}p<0.01$; $^{*}p<0.05$. This regression should be interpreted with strong caution as it is likely plagued by extensive omitted variable bias}
\end{tablenotes}
\end{threeparttable}
\caption{Statistical models}
\label{table:coefficients}
\end{center}
\end{table}
Which is rendered as:
Note that the custom note must start with \\item. It is also possible to have multiple items and/or use bullet points to format multiple notes like in a list:
texreg(lm(speed ~ dist, data = cars),
custom.note = paste("\\item[$\\bullet$] %stars.",
"\\item[$\\bullet$] This regression",
"should be interpreted with strong",
"caution as it is likely plagued by",
"extensive omitted variable bias."),
single.row = TRUE,
threeparttable = TRUE)
The formatting is not perfect as you cannot set the desired width of the table; the note just adjusts to the width of the respective table. But I think it should be less of a problem in a realistic usage scenario where more than one model is displayed at a time and some of the coefficient names are longer than in the example. This solution also supports longtable environments, in which case the threeparttablex package is used instead.
Here is an example of how you could make it look nice with two models:
fit <- lm(speed ~ dist, data = cars)
texreg(list(fit, fit),
custom.note = paste("\\item[\\hspace{-5mm}] %stars.",
"\\item[\\hspace{-5mm}] This regression",
"should be interpreted with strong",
"caution as it is likely plagued by",
"extensive omitted variable bias."),
single.row = TRUE,
threeparttable = TRUE)
This yields:
\begin{table}
\begin{center}
\begin{threeparttable}
\begin{tabular}{l c c}
\hline
& Model 1 & Model 2 \\
\hline
(Intercept) & $8.28 \; (0.87)^{***}$ & $8.28 \; (0.87)^{***}$ \\
dist & $0.17 \; (0.02)^{***}$ & $0.17 \; (0.02)^{***}$ \\
\hline
R$^2$ & $0.65$ & $0.65$ \\
Adj. R$^2$ & $0.64$ & $0.64$ \\
Num. obs. & $50$ & $50$ \\
\hline
\end{tabular}
\begin{tablenotes}[flushleft]
\scriptsize{\item[\hspace{-5mm}] $^{***}p<0.001$; $^{**}p<0.01$; $^{*}p<0.05$. \item[\hspace{-5mm}] This regression should be interpreted with strong caution as it is likely plagued by extensive omitted variable bias.}
\end{tablenotes}
\end{threeparttable}
\caption{Statistical models}
\label{table:coefficients}
\end{center}
\end{table}
Which renders as: