I have scatterplots of 2D data from two categories. I want to add density lines for each dimension -- not outside the plot (cf. Scatterplot with marginal histograms in ggplot2) but right on the plotting surface. I can get this for the x-axis dimension, like this:
set.seed(123)
dim1 <- c(rnorm(100, mean=1), rnorm(100, mean=4))
dim2 <- rnorm(200, mean=1)
cat <- factor(c(rep("a", 100), rep("b", 100)))
mydf <- data.frame(cbind(dim2, dim1, cat))
ggplot(data=mydf, aes(x=dim1, y=dim2, colour=as.factor(cat))) +
geom_point() +
stat_density(aes(x=dim1, y=(-2+(..scaled..))),
position="identity", geom="line")
It looks like this:
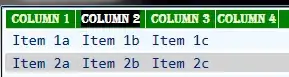
But I want an analogous pair of density curves running vertically, showing the distribution of points in the y-dimension. I tried
stat_density(aes(y=dim2, x=0+(..scaled..))), position="identity", geom="line)
but receive the error "stat_density requires the following missing aesthetics: x".
Any ideas? thanks