I have two data.frames:
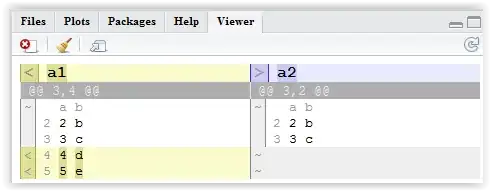
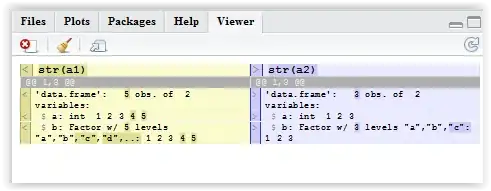
a1 <- data.frame(a = 1:5, b=letters[1:5])
a2 <- data.frame(a = 1:3, b=letters[1:3])
I want to find the rows a1 have that a2 doesn't.
Is there a built in function for this type of operation?
(p.s: I did write a solution for it, I am simply curious if someone already made a more crafted code)
Here is my solution:
a1 <- data.frame(a = 1:5, b=letters[1:5])
a2 <- data.frame(a = 1:3, b=letters[1:3])
rows.in.a1.that.are.not.in.a2 <- function(a1,a2)
{
a1.vec <- apply(a1, 1, paste, collapse = "")
a2.vec <- apply(a2, 1, paste, collapse = "")
a1.without.a2.rows <- a1[!a1.vec %in% a2.vec,]
return(a1.without.a2.rows)
}
rows.in.a1.that.are.not.in.a2(a1,a2)