Both SciPy and Numpy have built in functions for singular value decomposition (SVD). The commands are basically scipy.linalg.svd and numpy.linalg.svd. What is the difference between these two? Is any of them better than the other one?
- 1,757
- 5
- 22
- 41
-
3I don't know about the main behavior, but the `scipy` version has two additional options: 1) `overwrite_a`, which allows in-place modifications to the input and would reduce memory usage and possibly speed it up, and 2) `check_finite` which allows you to have the call assume the array is finite, saving some small overhead. – askewchan Sep 14 '15 at 16:58
3 Answers
From the FAQ page, it says scipy.linalg submodule provides a more complete wrapper for the Fortran LAPACK library whereas numpy.linalg tries to be able to build independent of LAPACK.
I did some benchmarks for the different implementation of the svd functions and found scipy.linalg.svd is faster than the numpy counterpart:
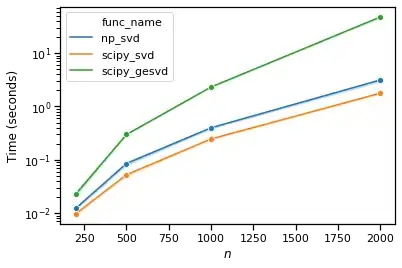
However, jax wrapped numpy, aka jax.numpy.linalg.svd is even faster:
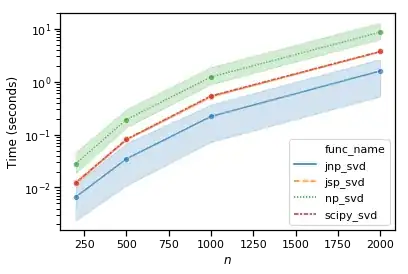
Full notebook for the benchmarks are available here.
- 1,294
- 13
- 22
-
-
These are somewhat moving targets. On both Windows and Linux, using either OpenBLAS or MKL, the performance of NumPy and SVD are now identical. JAX may still be faster, I did not test it. – Kevin S Aug 03 '20 at 17:08
-
Apart from the error checking, the actual work seems to be done within lapack
both with numpy and scipy.
Without having done any benchmarking, I guess the performance should be identical.
- 1,372
- 13
- 18
Another distinction is that np.linalg.svd can do vectorized svd calculations over large data arrays, where sp.linalg.svd will only do 1 at a time.
ex:
import numpy as np
import scipy as sp
data = np.random.random((3,3)) # a single matrix
data_array = np.random.random((10**6,3,3)) # one million matrices
# numpy svd
R,S,V = np.linalg.svd(data) # works
R,S,V = np.linalg.svd(data_array) # works
# scipy svd
R,S,V = sp.linalg.svd(data) # works
R,S,V = sp.linalg.svd(data_array) # fails !!!
I have not benchmarked this, but while a direct 1:1 comparison between the two might show sp.linalg.svd to be faster to compute, np.linalg.svd might be faster (or at least more convenient) when you need to compute the svd over a large data array.
- 5,365
- 4
- 31
- 48