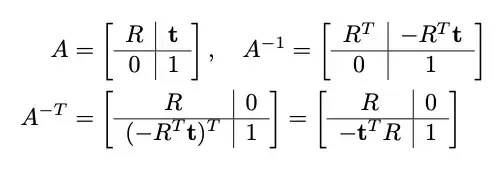First, don't be scared by the looks of this question ;)
I'm trying to implement a shape descriptor in matlab called Circular Blurred Shape Model, and part of this is to get a list of nearest neighbours for every radial segment as can be seen in Figure 1d)
I went for a straight and simple implementation in MATLAB but I'm stuck at Step 5 and 6 of the algorithm, mainly because I can't wrap my head around the definition:
Xb{c,s} = {b1, ..., b{c*s}} as the sorted set of the elements in B*
so that d(b*{c,s}, bi*) <= d(b*{c,s}, bj*), i<j
For me this sounds like a cascaded sorting, first sort by ascending distance and then by ascending index, but the nearest neighbours I find are not according to the paper.
As an example I show you the nearest neighbours I obtain for the segment b{4,1}, this is the one marked "EX" in Figure 1d)
I get the following list of nearest neighbors for b{4,1}: b{3,2}, b{3,1}, b{3,8}, b{2,1}, b{2,8}
correct according to the paper would be: b{4,2}, b{4,8}, b{3,2}, b{3,1}, b{3,8}
However my points actually are the closest set to the selected segment measured by euclidean distance! The distance b{4,1} <=> b{2,1} is smaller than b{4,1} <=> b{4,2} or b{4,1} <=> b{4,8}...
And here is my (ugly, but straight forward) MATLAB code:
width = 734;
height = 734;
assert(width == height, 'Image must be square in size!');
% Radius of the correlogram
R = width;
% Number of circles in correlogram
C = 4;
% Number of sections in correlogram
S = 8;
% "width" of ring segments
d = R/C;
% angle of one segment in degrees
g = 360/S;
% set of bins for the circular description of I
B = zeros(C, S);
% centroid coordinates for bins
B_star = zeros(C,S,2);
% calculate centroids of bins
for c=1:C
for s=1:S
alpha = deg2rad(max(s-1, 0)*g + g/2);
r = d*max((c-1),0) + d/2;
B_star(c,s,1) = r*cos(alpha);
B_star(c,s,2) = r*sin(alpha);
end
end
% create sorted list of bin numbers which fullfill
% d(b{c,s}*, bi*) <= d(b{c,s}, bj*) where i<j
% B_star_dists is a simple square distance matrix for getting
% the distance between two centroids c_i,s_i and c_j,s_j
B_star_dists = zeros(C*S, C*S);
for i=1:C*S
[c_i, s_i] = ind2sub([C,S], i);
% x,y centroid coordinates for point i
b_star_i = [B_star(c_i, s_i, 1), B_star(c_i, s_i, 2)];
for j=1:C*S
[c_j, s_j] = ind2sub([C,S], j);
% x,y centroid coordinates for point j
b_star_j = [B_star(c_j, s_j, 1), B_star(c_j, s_j, 2)];
% store the euclidean distance between these two centroids
% in the distance matrix.
B_star_dists(i,j) = norm(b_star_i - b_star_j);
end
end
% calculate nearest neighbour "centroids" for each centroid
% B_NN is a cell array, B{idx} gives an array of indexes to the
% nearest neighbour centroids.
B_NN = cell(C*S, 1);
for i=1:C*S
[c_i, s_i] = ind2sub([C,S], i);
% get a (C*S)x2 matrix of all distances, the first column are the array
% indexes and the second column are the distances e.g
% 1 d1
% 2 d2
% .. ..
% CS d{c,s}
dists = [transpose(1:C*S), B_star_dists(:, i)];
% sort ascending by the distances first (e.g second column) then
% sort ascending by the array index (e.g first column)
dists = sortrows(dists, [2,1]);
% middle section has nine neighbours, set as default
neighbour_count = 9;
if c_i == 1
% inner region has S+3 neighbours
neighbour_count = S+3;
elseif c_i == C
% outer most ring has 6 neighbours
neighbour_count = 6;
end
B_NN{i} = dists(1:neighbour_count,1);
end
% FROM HERE ON JUST VISUALIZATION CODE
figure(1);
hold on;
for c=1:C
% plot circles
r = c*d;
plot(r*cos(0:pi/50:2*pi), r*sin(0:pi/50:2*pi), 'k:');
end
for s=1:S
% plot lines
line_len = C*d;
alpha = deg2rad(s*g);
start_pt = [0, 0];
end_pt = start_pt + line_len.*[cos(alpha), sin(alpha)];
plot([start_pt(1), end_pt(1)], [start_pt(2), end_pt(2)], 'k-');
end
for c=1:C
% plot centroids of segments
for s=1:S
segment_centroid = B_star(c,s, :);
plot(segment_centroid(1), segment_centroid(2), '.k');
end
end
% plot some nearest neighbours
% list of [C;S]
plot_nn = [4;1];
for i = 1:size(plot_nn,2)
start_c = plot_nn(1,i);
start_s = plot_nn(2,i);
start_pt = [B_star(start_c, start_s,1), B_star(start_c, start_s,2)];
start_idx = sub2ind([C, S], start_c, start_s);
plot(start_pt(1), start_pt(2), 'xb');
nn_idx_list = B_NN{start_idx};
for j = 1:length(nn_idx_list)
nn_idx = nn_idx_list(j);
[nn_c, nn_s] = ind2sub([C, S], nn_idx);
nn_pt = [B_star(nn_c, nn_s,1), B_star(nn_c, nn_s,2)];
plot(nn_pt(1), nn_pt(2), 'xr');
end
end
The full paper can be found here