I'm printing two plots with ggplot into an R Markdown HTML output, but I'd like them to appear side by side. Is this possible? Could I set the size of the figures as well?
SO far I can only print them one after the other. I also tried the multiplot function from the R Cookbook, but that severely distorts the plots...
Thanks!
title: "HT Chip MiSeq/HiSeq Analysis"
date: "October 1, 2015"
output:
html_document:
highlight: haddock
theme: flatly
---
```{r plots, echo=FALSE}
genesDetectedDensity_MiSeq <- ggplot(meta.miseq) + geom_density(aes(genesDetected, fill=column, color=seqRun), alpha=0.2) + scale_x_continuous(limits=c(0,2000), breaks=seq(0, 2000, 100)) + ggtitle("Genes Detected across cells from MiSeq Runs")
return(genesDetectedDensity_MiSeq)
genesDetectedHistogram_MiSeq <- ggplot(meta.miseq) + geom_bar(aes(genesDetected, fill=column, color=seqRun), position="dodge", binwidth=50, alpha=0.2) + scale_x_continuous(limits=c(0,2000), breaks=seq(0, 2000, 100)) + ggtitle("Genes Detected across cells from MiSeq Runs")
return(genesDetectedHistogram_MiSeq)
```
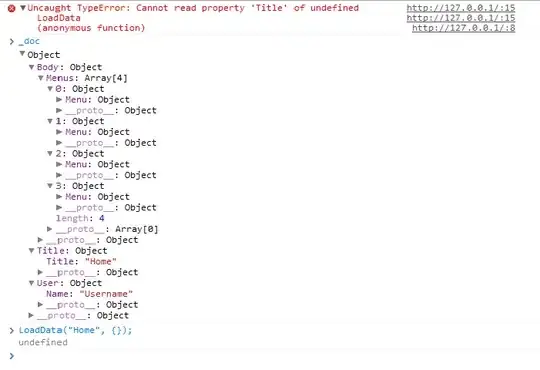
This produces the following:
UPDATE: Following the suggestion I received below, I tried using the gridExtra library, and printed the plots by adding:
grid.arrange(genesDetectedDensity_MiSeq, genesDetectedHistogram_MiSeq, ncol=2)
This almost works, but it's still kind of messy: