Genetic algorithms are mocking biological processes where chromosomes crossover at one point and exchange their parts after the crossover point if we are talking about single point crossover.
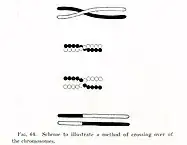
As you can see in picture above parents exchange "tail" parts of the chromosome after the crossover point. Therefore you have only 2 offspring/children produced by crossover. That's how crossover occurs in nature, how biologists describe it.
If you refer to any literature dealing with topic of Genetic Algorithms they also state this convention that when using single point crossover parent chromosomes are split into head and tail denoted as H/T like that (see citation below):
H T
123456.789
H T
ABCDEF.GHI
Therefore offspring produced with this crossover will be:
123456GHI
ABCDEF789
Following this convention is much better than creating all possible combinations and then selecting random or fittest of the offspring as it is computationally more efficient. If you want to solve more complex problems you just simply increase size of the population to allow more diversity.
"Single point crossover: A single random cut is made, producing two head sections and two tail sections. The two tail sections are then swapped to produce
two new individuals (chromosomes)".
Genetic Algorithms and Genetic Programming:
Affenzeller, Michael
Wagner, Stefan
Winkler, Stephan,
ISBN: 1584886293
Alternatively you can use multipoint crossover which follows similar convention where chromosomes are split into sections and parents exchange parts in a way that offspring chromosome is just alternation of parents chromosomes so if you have parents with chromosomes:
A1.A2.A3.A4
B1.B2.B3.B4
|
| this will produce offspring
|
A1 B2 A3 B4
and
B1 A2 B3 A4
This answer might help you as well:
Crossover of chromosomes with different length
