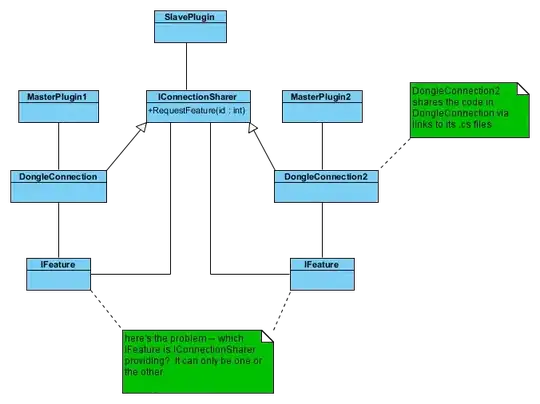
I'm working with a data frame of size 2 x 400. I need to graph this (let's call it data set A) on the same graph as the main data set for my project.
All I need is the general shape of data set A's graph. ie i only need to see the trend.
The scale that data set A takes place on happens to be much smaller than that of the main graph. So dataset A just looks like a horizontal line.
I decided to scale data set A by multiplying it by a factor of... I tried various values to get the optimum vertical scaling, which leads me to the problem I'm having.
When trying to find the ideal multiplicative factor by trial and error, I expected the general shape of data set A's graph to retain its shape, and only vary in its relative vertical points . ie the horizontal coordinates of all maxes and mins shouldn't move, and only the vertical points should be moving. but this wasn't happening. I'd like to know why.
Here's the data set A (yellow), when multiplied by factor of 3:
factor of 5:
The yellow dots are the geom_point and the yellow curve is the corresponding geom_smooth.
EDIT: here is my the code original code: I haven't had much formal training with code. I'm apologize for any messiness!
library("ggplot2")
library("dplyr")
# READ IN DATA
temp_data <-read.table(col.names = "y",
"C:/Users/Ben/Documents/Visual Studio 2013/Projects/Home/Home/steamdata2.txt")
boilpoint <- which(temp_data$y == "boil") # JUST A MARKER..
temp_data <- filter(temp_data, y != "boil") # GETTING RID OF THE MARKER ENTRY
# DON'T KNOW WHY BUT I HAD TO DO THIS INTERMEDIATE STEP
# BEFORE I COULD CONVERT FROM FACTOR -> NUMERIC
temp_data$y <- as.character(temp_data$y)
# CONVERTING TO NUMERIC
temp_data$y <- as.numeric(temp_data$y)
# GETTING RID OF BASICALLY THE LAST ENTRY WHICH HAS THE LARGEST VALUE
temp_data <- filter(temp_data, y<max(temp_data$y))
# ADD ANOTHER COLUMN WITH THE ROW NUMBER,
# BECAUSE I DON'T KNOW HOW TO ACCESS THIS FOR GGPLOT
temp_data <- transform(temp_data, x = 1:nrow(temp_data))
n <- nrow(temp_data) # Num of readings
period <- temp_data[n,1] # (sec)
RpS <- n / period # Avg Readings per Second
MIN <- min(temp_data$y)
MAX <- max(temp_data$y)
# DERIVATIVE OF ORIGINAL
deriv <- data.frame(matrix(ncol=2, nrow=n))
# ADD ANOTHER COLUMN TO ACCESS ROW NUMBERS FOR GGPLOT LATER
colnames(deriv) <- c("y","x")
deriv <- transform(deriv, x = c(1:n))
# FILL DERIVATIVE DATAFRAME
deriv[1, 1] <- 0
for(i in 2:n){
deriv[i - 1, 1] <- temp_data[i, 1] - temp_data[i - 1, 1]
}
deriv <- filter(deriv, y != 0)
# DID THE SAME FOR SECOND DERIVATIVE
dderiv <- data.frame(matrix(ncol = 2, nrow = nrow(deriv)))
colnames(dderiv) <- c("y", "x")
dderiv <- transform(dderiv, x=rep(0, nrow(deriv)))
dderiv[1, 1] <- 0
for(i in 2:nrow(deriv)) {
dderiv$y[i - 1] <- (deriv$y[i] - deriv$y[i - 1]) /
(deriv$x[i] - deriv$x[i - 1])
dderiv$x[i - 1] <- deriv$x[i] + (deriv$x[i] - deriv$x[i - 1]) / 2
}
dderiv <- filter(dderiv, y!=0)
# HERE'S WHERE I FACTOR BY VARIOUS MULTIPLES
deriv <- MIN + deriv * 3
dderiv <- MIN + dderiv * 3
graph <- ggplot(temp_data, aes(x, y)) + geom_smooth()
graph <- graph + geom_point(data = deriv, color = "yellow")
graph <- graph + geom_smooth(data = deriv, color = "yellow")
graph <- graph + geom_point(data = dderiv, color = "green")
graph <- graph + geom_smooth(data = dderiv, color = "green")
graph <- graph + geom_vline(xintercept = boilpoint, color = "red")
graph <- graph + xlab("Readings (n)") +
ylab(expression(paste("Temperature (",degree,"C)")))
graph <- graph + xlim(c(0,n)) + ylim(c(MIN, MAX))