Im using the varImp function of the caret package and Im trying to plot the resulting dataframe that it creates. Here is the code:
RocImp2 <- varImp(svmFit, scale = FALSE)
and Im using a simple plot to plot the chart:
plot(RocImp2)
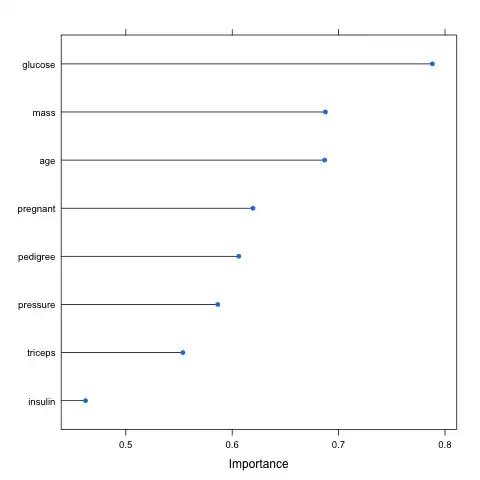
The results are supposed to look like this:
whereas mine looks like this:
Here is a copy of the dataframe:
disregard the names in the plot as they are from another example.