I am trying to use Python and scipy.integrate.odeint to simulate the following dynamical system:
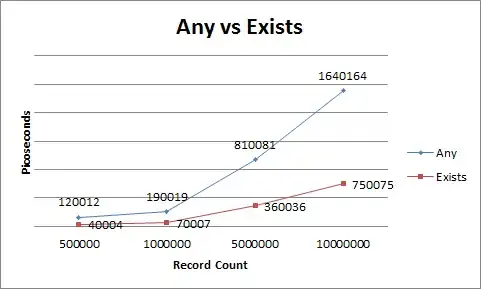
But this integration breaks numerically in Python resulting in the following and similar images (usually even worse than this):
Generated using the following in iPython/Jupyter notebook:
import numpy as np
from scipy.integrate import odeint
import matplotlib.pyplot as plt
%matplotlib inline
f = lambda x,t: -np.sign(x)
x0 = 3
ts = np.linspace(0,10,1000)
xs = odeint(f,x0,ts)
plt.plot(ts,xs)
plt.show()
Any advice how to better simulate such a system with discontinuous dynamics?
Edit #1:
Example result when ran with smaller timestep, ts = np.linspace(0,10,1000000), in response to @Hun's answer. This is also an incorrect result according to my expectations.