I'm plotting data using the matplotlib functions pcolormesh and imshow and when I use pcolormesh it produces artifacts where it seems to slide some of the data around:
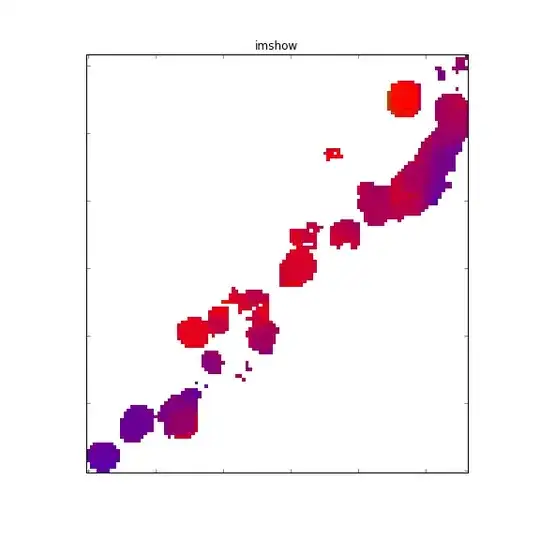
whereas imshow does not:
I was able to produce an example that has the same artifacting
import numpy as np
import pandas as pd
data = pd.DataFrame({'x':np.random.normal(loc=0.5, size=5000)
, 'y':np.random.normal(loc=0.5, size=5000)
, 'z':np.random.normal(loc=0.5, size=5000)})
data_pivot = data.pivot(index='x', columns='y', values='z')
x = data_pivot.index.values
y = data_pivot.columns.values
z = data_pivot.values
masked_data = np.ma.masked_invalid(z)
which produces the following figures like so:
fig, ax = plt.subplots(1, figsize=(8,8))
ax.pcolormesh(x, y, masked_data)
Where do these artifacts come from? There isn't anything wrong with the data as far as I can tell since the original data and the made up data produce the same result.