So. We have a messy data stored in a TSV file that I need to analyse. This is how it looks
status=200 protocol=http region_name=Podolsk datetime=2016-03-10 15:51:58 user_ip=0.120.81.243 user_agent=Mozilla/5.0 (Windows NT 6.1; WOW64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/48.0.2564.116 Safari/537.36 user_id=7885299833141807155 user_vhost=tindex.ru method=GET page=/search/
And the problem is that some of the rows have different column order / some of them missing values and I need to get rid of that with high performance (since the datasets I am working with are up to 100 Gigabytes).
Data = pd.read_table('data/data.tsv', sep='\t+',header=None,names=['status', 'protocol',\
'region_name', 'datetime',\
'user_ip', 'user_agent',\
'user_id', 'user_vhost',\
'method', 'page'], engine='python')
Clean_Data = (Data.dropna()).reset_index(drop=True)
Now I got rid of missing values but one problem still remains!
This is how the data looks:
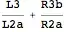
And this is how the problem looks:
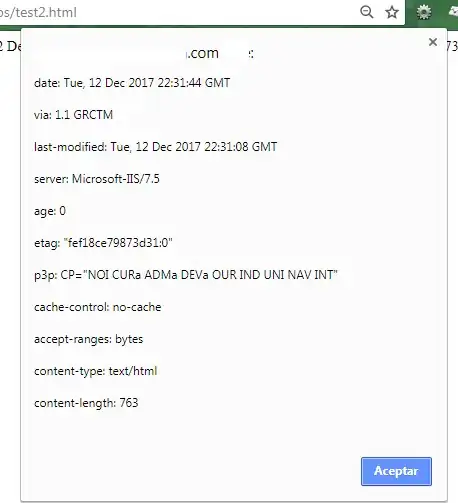
As you can see some of columns are offset. I made a very low-performance solution
ids = Clean_Data.index.tolist()
for column in Clean_Data.columns:
for row, i in zip(Clean_Data[column], ids):
if np.logical_not(str(column) in row):
Clean_Data.drop([i], inplace=True)
ids.remove(i)
So now the data looks good... at least I can work with it! But what is the High-Performance ALTERNATIVE to the method I made above?
Update on unutbu code: traceback error
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
<ipython-input-4-52c9d76f9744> in <module>()
8 df.index.names = ['index', 'num']
9
---> 10 df = df.set_index('field', append=True)
11 df.index = df.index.droplevel(level='num')
12 df = df['value'].unstack(level=1)
/Users/Peter/anaconda/lib/python2.7/site-packages/pandas/core/frame.pyc in set_index(self, keys, drop, append, inplace, verify_integrity)
2805 if isinstance(self.index, MultiIndex):
2806 for i in range(self.index.nlevels):
-> 2807 arrays.append(self.index.get_level_values(i))
2808 else:
2809 arrays.append(self.index)
/Users/Peter/anaconda/lib/python2.7/site-packages/pandas/indexes/multi.pyc in get_level_values(self, level)
664 values = _simple_new(filled, self.names[num],
665 freq=getattr(unique, 'freq', None),
--> 666 tz=getattr(unique, 'tz', None))
667 return values
668
/Users/Peter/anaconda/lib/python2.7/site-packages/pandas/indexes/range.pyc in _simple_new(cls, start, stop, step, name, dtype, **kwargs)
124 return RangeIndex(start, stop, step, name=name, **kwargs)
125 except TypeError:
--> 126 return Index(start, stop, step, name=name, **kwargs)
127
128 result._start = start
/Users/Peter/anaconda/lib/python2.7/site-packages/pandas/indexes/base.pyc in __new__(cls, data, dtype, copy, name, fastpath, tupleize_cols, **kwargs)
212 if issubclass(data.dtype.type, np.integer):
213 from .numeric import Int64Index
--> 214 return Int64Index(data, copy=copy, dtype=dtype, name=name)
215 elif issubclass(data.dtype.type, np.floating):
216 from .numeric import Float64Index
/Users/Peter/anaconda/lib/python2.7/site-packages/pandas/indexes/numeric.pyc in __new__(cls, data, dtype, copy, name, fastpath, **kwargs)
105 # with a platform int
106 if (dtype is None or
--> 107 not issubclass(np.dtype(dtype).type, np.integer)):
108 dtype = np.int64
109
TypeError: data type "index" not understood
Pandas version : 0.18.0-np110py27_0
Update
Everything worked... Thanks everybody!