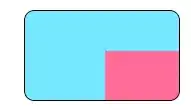
I'm looking for a way to produce a diagonal slash from the bottom left the to top right corner of a cell within a plot made using geom_tile.
The input is a melted data frame with two categorical factor columns, sample and gene. I'd like to use something like geom_segment, but I'm not able to specify fractional increments. Any ideas on the best way to accomplish this?
edit: Here is a reproducible example, I can't share one from my own data, as it's protected patient information.
df <- data_frame( gene = c('TP53','TP53','MTOR','BRACA1'),
sample = c('A','B','A','B'),
diagonal = c(FALSE,TRUE,TRUE,FALSE),
effect = c('missense', 'nonsense', 'missense', 'silent') )
ggplot(df, aes(sample, gene)) + geom_tile(aes(fill = effect))
what I'm looking for: