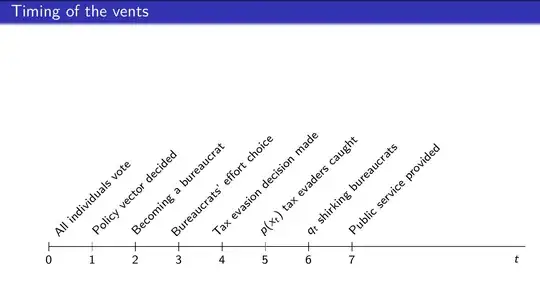
I was looking for a similar (but slightly different) solution. Posting here in case it's useful to anyone else.
In my case, I needed a more general solution that allows each letter to be repeated an arbitrary number of times. Here's what I came up with:
library(tidyverse)
df <- data.frame(letters = letters[1:4])
df
> df
letters
1 a
2 b
3 c
4 d
Let's say I want 2 A's, 3 B's, 2 C's and 4 D's:
df %>%
mutate(count = c(2, 3, 2, 4)) %>%
group_by(letters) %>%
expand(count = seq(1:count))
# A tibble: 11 x 2
# Groups: letters [4]
letters count
<fctr> <int>
1 a 1
2 a 2
3 b 1
4 b 2
5 b 3
6 c 1
7 c 2
8 d 1
9 d 2
10 d 3
11 d 4
If you don't want to keep the count column:
df %>%
mutate(count = c(2, 3, 2, 4)) %>%
group_by(letters) %>%
expand(count = seq(1:count)) %>%
select(letters)
# A tibble: 11 x 1
# Groups: letters [4]
letters
<fctr>
1 a
2 a
3 b
4 b
5 b
6 c
7 c
8 d
9 d
10 d
11 d
If you want the count to reflect the number of times each letter is repeated:
df %>%
mutate(count = c(2, 3, 2, 4)) %>%
group_by(letters) %>%
expand(count = seq(1:count)) %>%
mutate(count = max(count))
# A tibble: 11 x 2
# Groups: letters [4]
letters count
<fctr> <dbl>
1 a 2
2 a 2
3 b 3
4 b 3
5 b 3
6 c 2
7 c 2
8 d 4
9 d 4
10 d 4
11 d 4