I am finding it hard to link the theory with the implementation. I would appreciate help in knowing where my understanding is wrong.
Notations - matrix in bold capital and vectors in bold font small letter
 is a dataset on
is a dataset on  observations, each of
observations, each of  variables. So, given these observed
variables. So, given these observed  -dimensional data vectors, the
-dimensional data vectors, the 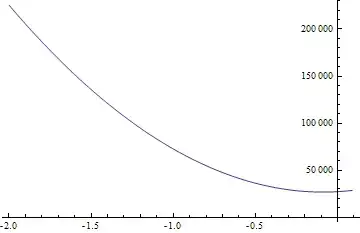 -dimensional principal axes are
-dimensional principal axes are  , for
, for  in
in  where
where  is the target dimension.
is the target dimension.
The 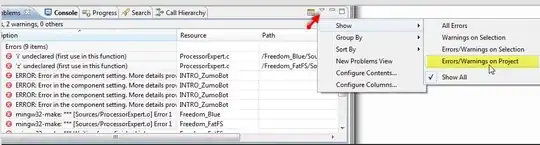 principal components of the observed data matrix would be
principal components of the observed data matrix would be  where matrix
where matrix  , matrix
, matrix  , and matrix
, and matrix  .
.
Columns of  form an orthogonal basis for the
form an orthogonal basis for the  features and the output
features and the output 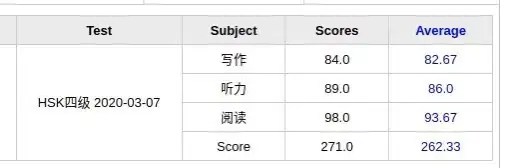 is the principal component projection that minimizes the squared reconstruction error:
is the principal component projection that minimizes the squared reconstruction error:
and the optimal reconstruction of  is given by
is given by  .
.
The data model is
X(i,j) = A(i,:)*S(:,j) + noise
where PCA should be done on X to get the output S. S must be equal to Y.
Problem 1: The reduced data Y is not equal to S that is used in the model. Where is my understanding wrong?
Problem 2: How to reconstruct such that the error is minimum?
Please help. Thank you.
clear all
clc
n1 = 5; %d dimension
n2 = 500; % number of examples
ncomp = 2; % target reduced dimension
%Generating data according to the model
% X(i,j) = A(i,:)*S(:,j) + noise
Ar = orth(randn(n1,ncomp))*diag(ncomp:-1:1);
T = 1:n2;
%generating synthetic data from a dynamical model
S = [ exp(-T/150).*cos( 2*pi*T/50 )
exp(-T/150).*sin( 2*pi*T/50 ) ];
% Normalizing to zero mean and unit variance
S = ( S - repmat( mean(S,2), 1, n2 ) );
S = S ./ repmat( sqrt( mean( Sr.^2, 2 ) ), 1, n2 );
Xr = Ar * S;
Xrnoise = Xr + 0.2 * randn(n1,n2);
h1 = tsplot(S);
X = Xrnoise;
XX = X';
[pc, ~] = eigs(cov(XX), ncomp);
Y = XX*pc;
UPDATE [10 Aug]
Based on the Answer, here is the full code that
clear all
clc
n1 = 5; %d dimension
n2 = 500; % number of examples
ncomp = 2; % target reduced dimension
%Generating data according to the model
% X(i,j) = A(i,:)*S(:,j) + noise
Ar = orth(randn(n1,ncomp))*diag(ncomp:-1:1);
T = 1:n2;
%generating synthetic data from a dynamical model
S = [ exp(-T/150).*cos( 2*pi*T/50 )
exp(-T/150).*sin( 2*pi*T/50 ) ];
% Normalizing to zero mean and unit variance
S = ( S - repmat( mean(S,2), 1, n2 ) );
S = S ./ repmat( sqrt( mean( S.^2, 2 ) ), 1, n2 );
Xr = Ar * S;
Xrnoise = Xr + 0.2 * randn(n1,n2);
X = Xrnoise;
XX = X';
[pc, ~] = eigs(cov(XX), ncomp);
Y = XX*pc; %Y are the principal components of X'
%what you call pc is misleading, these are not the principal components
%These Y columns are orthogonal, and should span the same space
%as S approximatively indeed (not exactly, since you introduced noise).
%If you want to reconstruct
%the original data can be retrieved by projecting
%the principal components back on the original space like this:
Xrnoise_reconstructed = Y*pc';
%Then, you still need to project it through
%to the S space, if you want to reconstruct S
S_reconstruct = Ar'*Xrnoise_reconstructed';
plot(1:length(S_reconstruct),S_reconstruct,'r')
hold on
plot(1:length(S),S)
The plot is 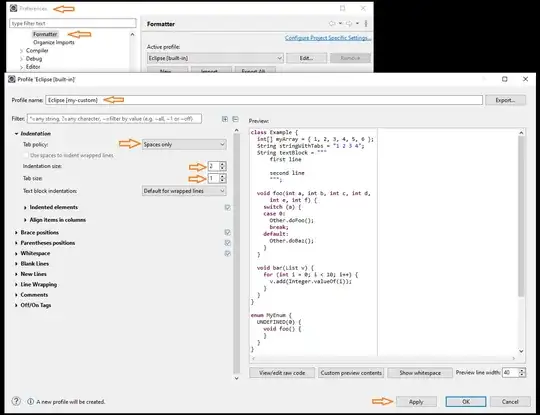 which is very different from the one that is shown in the Answer. Only one component of S exactly matches with that of S_reconstructed. Shouldn't the entire original 2 dimensional space of the source input S be reconstructed?
Even if I cut off the noise, then also onely one component of S is exactly reconstructed.
which is very different from the one that is shown in the Answer. Only one component of S exactly matches with that of S_reconstructed. Shouldn't the entire original 2 dimensional space of the source input S be reconstructed?
Even if I cut off the noise, then also onely one component of S is exactly reconstructed.