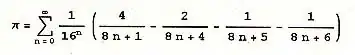What follows is an automated global method for isolating an area of low color saturation (e.g. a b/w page) against a colored background. This may work well as an alternative approach when other approaches based on adaptive thresholding of grayscale-converted images fail.
First, we load an RGB image I, covert from RGB to HSV, and isolate the saturation channel:
I = imread('/path/to/image.jpg');
Ihsv = rgb2hsv(I); % convert image to HSV
Isat = Ihsv(:,:,2); % keep only saturation channel
In general, a good first step when deciding how to proceed with any object detection task is to examine the distribution of pixel values. In this case, these values represent the color saturation levels at each point in our image:
% Visualize saturation value distribution
imhist(Isat); box off
From this histogram, we can see that there appear to be at least 3 distinct peaks. Given that our target is a black and white sheet of paper, we’re looking to isolate saturation values at the lower end of the spectrum. This means we want to find a threshold that separates the lower 1-2 peaks from the higher values.
One way to do this in an automated way is through Gaussian Mixture Modeling (GMM). GMM can be slow, but since you’re processing images offline I assume this is not an issue. We’ll use Matlab’s fitgmdist function here and attempt to fit 3 Gaussians to the saturation image:
% Find threshold for calling ROI using GMM
n_gauss = 3; % number of Gaussians to fit
gmm_opt = statset('MaxIter', 1e3); % max iterations to converge
gmmf = fitgmdist(Isat(:), n_gauss, 'Options', gmm_opt);
Next, we use the GMM fit to classify each pixel and visualize the results of our GMM classification:
% Classify pixels using GMM
gmm_class = cluster(gmmf, Isat(:));
% Plot histogram, colored by class
hold on
bin_edges = linspace(0,1,256);
for j=1:n_gauss, histogram(Isat(gmm_class==j), bin_edges); end
In this example, we can see that the GMM ended up grouping the 2 far left peaks together (blue class) and split the higher values into two classes (yellow and red). Note: your colors might be different, since GMM is sensitive to random initial conditions. For our use here, this is probably fine, but we can check that the blue class does in fact capture the object we’d like to isolate by visualizing the image, with pixels colored by class:
% Visualize classes as image
im_class = reshape(gmm_class ,size(Isat));
imagesc(im_class); axis image off

So it seems like our GMM segmentation on saturation values gets us in the right ballpark - grouping the document pixels (blue) together. But notice that we still have two problems to fix. First, the big bar across the bottom is also included in the same class with the document. Second, the text printed on the page is not being included in the document class. But don't worry, we can fix these problems by applying some filters on the GMM-grouped image.
First, we’ll isolate the class we want, then do some morphological operations to low-pass filter and fill gaps in the objects.
Isat_bw = im_class == find(gmmf.mu == min(gmmf.mu)); %isolate desired class
opened = imopen(Isat_bw, strel('disk',3)); % morph open
closed = imclose(Isat_bw, strel('disk',50)); % morph close
imshow(closed)
Next, we’ll use a size filter to isolate the document ROI from the big object at the bottom. I’ll assume that your document will never fill the entire width of the image and that any solid objects bigger than the sheet of paper are not wanted. We can use the regionprops function to give us statistics about the objects we detect and, in this case, we’ll just return the objects’ major axis length and corresponding pixels:
% Size filtering
props = regionprops(closed,'PixelIdxList','MajorAxisLength');
[~,ridx] = min([props.MajorAxisLength]);
output_im = zeros(numel(closed),1);
output_im(props(ridx).PixelIdxList) = 1;
output_im = reshape(output_im, size(closed));
% Display final mask
imshow(output_im)
Finally, we are left with output_im - a binary mask for a single solid object corresponding to the document. If this particular size filtering rule doesn’t work well on your other images, it should be possible to find a set of values for other features reported by regionprops (e.g. total area, minor axis length, etc.) that give reliable results.
A side-by-side comparison of the original and the final masked image shows that this approach produces pretty good results for your sample image, but some of the parameters (like the size exclusion rules) may need to be tuned if results for other images aren't quite as nice.
% Display final image
image([I I.*uint8(output_im)]); axis image; axis off
One final note: be aware that the GMM algorithm is sensitive to random initial conditions, and therefore might randomly fail, or produce undesirable results. Because of this, it's important to have some kind of quality control measures in place to ensure that these random failures are detected. One possibility is to use the posterior probabilities of the GMM model to form some kind of criteria for rejecting a certain fit, but that’s beyond the scope of this answer.