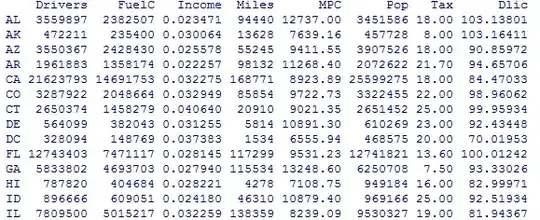
I want to add some information to my tree. Let's say for instance I have a database like this :
library(rpart)
library(rpart.plot)
set.seed(1)
mydb<-data.frame(results=rnorm(1000,0,1),expo=runif(1000),var1=sample(LETTERS[1:4],1000,replace=T),
var2=sample(LETTERS[5:6],1000,replace=T),var3=sample(LETTERS[20:25], 1000,replace=T))
I can run a tree :
mytree<-rpart(results~var1+var2+var3,data=mydb,cp=0)
pfit<- prune(mytree, cp=mytree$cptable[4,"CP"])
prp(pfit,type=1,extra=100,fallen.leaves=F,shadow.col="darkgray",box.col=rgb(0.8,0.9,0.8))
And it's ok for me, but let's imagine I want to know the average exposure for each leaf.
I know i can add some informations to prp, for instance the weight of each leaf with a function :
node.fun1 <- function(x, labs, digits, varlen)
{
paste("Weight \n",x$frame$wt)
}
prp(pfit,type=1,extra=100,fallen.leaves=F,shadow.col="darkgray",box.col=rgb(0.8,0.9,0.8),node.fun = node.fun1)
But it works only if it's calculated in frame, the results of the rpart function.
My question :
How can I add custom informations to the plot, like the average exposure, or any other function that calculates custom indicators and add it to the table frame ?