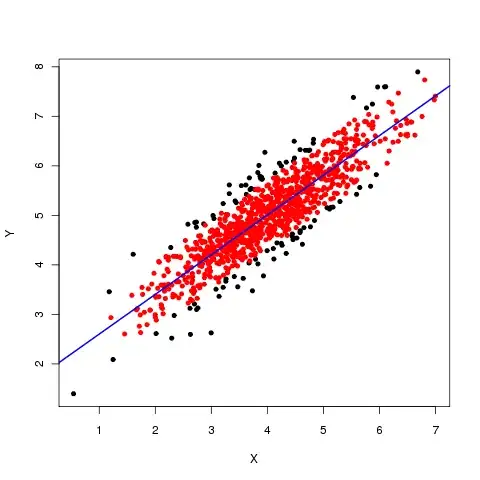
In general it is very difficult to see the effect of a noise filter by eye in a 2D plot.
Let me demonstrate this by example. Suppose we have this noisy picture:

Let me now convert this image to a 3d mesh plot. Then it will look like this. The noise becomes very clear but also the differences in depth between the left and right side of the picture.

The library findpeaks contains many filters which are utilized from various (old python 2) libraries and rewritten to python 3. Applying the filters is very easy as shown below. Note that this example seems not very representative for a SAR image as there is no speckle noise. A mean or median filter seems to perform very well in this example. In speckle noise images where local heights are important, such mean/median filters can remove the peaks and thus destroy the signal of interest.
Install by:
pip install findpeaks
Run by:
from findpeaks import findpeaks
# Read image
img = cv2.imread('noise.png')
filters = [None, 'lee','lee_enhanced','kuan', 'fastnl','bilateral','frost','median','mean']
for getfilter in filters:
fp = findpeaks(method='topology', scale=False, denoise=getfilter, togray=True, imsize=False, window=15)
fp.fit(img)
fp.plot_mesh(wireframe=False, title=str(getfilter), view=(30,30))
If you directly want to use the denoising filters, it can be done as following:
import findpeaks
import matplotlib.pyplot as plt
# Read image
img = cv2.imread('noise.png')
# filters parameters
# window size
winsize = 15
# damping factor for frost
k_value1 = 2.0
# damping factor for lee enhanced
k_value2 = 1.0
# coefficient of variation of noise
cu_value = 0.25
# coefficient of variation for lee enhanced of noise
cu_lee_enhanced = 0.523
# max coefficient of variation for lee enhanced
cmax_value = 1.73
# Some pre-processing
# Make grey image
img = findpeaks.stats.togray(img)
# Scale between [0-255]
img = findpeaks.stats.scale(img)
# Denoising
# fastnl
img_fastnl = findpeaks.stats.denoise(img, method='fastnl', window=winsize)
# bilateral
img_bilateral = findpeaks.stats.denoise(img, method='bilateral', window=winsize)
# frost filter
image_frost = findpeaks.frost_filter(img, damping_factor=k_value1, win_size=winsize)
# kuan filter
image_kuan = findpeaks.kuan_filter(img, win_size=winsize, cu=cu_value)
# lee filter
image_lee = findpeaks.lee_filter(img, win_size=winsize, cu=cu_value)
# lee enhanced filter
image_lee_enhanced = findpeaks.lee_enhanced_filter(img, win_size=winsize, k=k_value2, cu=cu_lee_enhanced, cmax=cmax_value)
# mean filter
image_mean = findpeaks.mean_filter(img, win_size=winsize)
# median filter
image_median = findpeaks.median_filter(img, win_size=winsize)
plt.figure(); plt.imshow(img_fastnl, cmap='gray'); plt.title('Fastnl')
plt.figure(); plt.imshow(img_bilateral, cmap='gray'); plt.title('Bilateral')
plt.figure(); plt.imshow(image_frost, cmap='gray'); plt.title('Frost')
plt.figure(); plt.imshow(image_kuan, cmap='gray'); plt.title('Kuan')
plt.figure(); plt.imshow(image_lee, cmap='gray'); plt.title('Lee')
plt.figure(); plt.imshow(image_lee_enhanced, cmap='gray'); plt.title('Lee Enhanced')
plt.figure(); plt.imshow(image_mean, cmap='gray'); plt.title('Mean')
plt.figure(); plt.imshow(image_median, cmap='gray'); plt.title('Median')
If you want to play around with the library, more examples can be found over here.