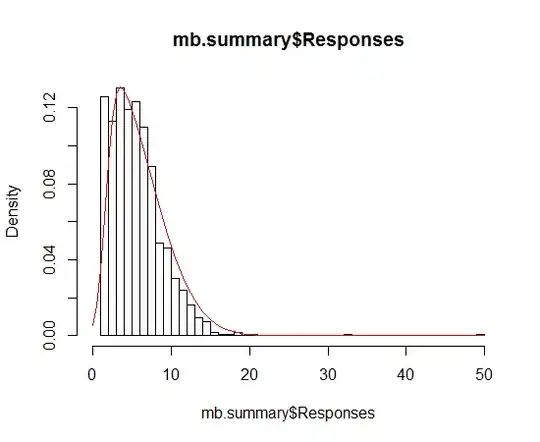
I am trying to get a picture like this:
In the picture, the parameters (a, b, and c of the triangle distribution ), their distributions, and confidence intervals of the parameters are based on the original datasets, and simulated ones are generated by parametric and nonparametric bootstrap. How to draw a picture like this in R? Can you give a simple example like this? Thank you very much!
Here is my code.
x1<-c(1300,541,441,35,278,167,276,159,126,170,251.3,155.84,187.01,850)
x2<-c(694,901,25,500,42,2.2,7.86,50)
x3<-c(2800,66.5,420,260,50,370,17)
x4<-c(12,3.9,10,28,84,138,6.65)
y1<-log10(x1)
y2<-log10(x2)
y3<-log10(x3)
y4<-log10(x4)
#Part 1 (Input the data) In this part, I have calculated the parameters (a and b) and the confidence interval (a and b ) by MLE and PB-MLE with different data sets(x1 to x4)
#To calculate the parameters (a and b) with data sets x1
y.n<-length(y1)
y.location<-mean(y1)
y.var<-(y.n-1)/y.n*var(y1)
y.scale<-sqrt(3*y.var)/pi
library(stats4)
ll.logis<-function(location=y.location,scale=y.scale){-sum(dlogis(y1,location,scale,log=TRUE))}
fit.mle<-mle(ll.logis,method="Nelder-Mead")
a1_mle<-coef(fit.mle)[1]
b1_mle<-coef(fit.mle)[2]
summary(a1_mle)# To calculate the parameters (a)
summary(b1_mle)# To calculate the parameters (b)
confint(fit.mle)# To calculate the confidence interval (a and b ) by MLE
# load fitdistrplus package for using fitdist function
library(fitdistrplus)
# fit logistic distribution using MLE method
x1.logis <- fitdist(y1, "logis", method="mle")
A<- bootdist(x1.logis, bootmethod="param", niter=1001)
summary(A) # To calculate the parameters (a and b ) and the confidence interval (a and b ) by parametric bootstrap
a <- A$estim
a1<-c(a$location)
b1<-c(a$scale)
#To calculate the parameters (a and b) with data sets x2
y.n<-length(y2)
y.location<-mean(y2)
y.var<-(y.n-1)/y.n*var(y2)
y.scale<-sqrt(3*y.var)/pi
library(stats4)
ll.logis<-function(location=y.location,scale=y.scale){-sum(dlogis(y2,location,scale,log=TRUE))}
fit.mle<-mle(ll.logis,method="Nelder-Mead")
a2_mle<-coef(fit.mle)[1]
b2_mle<-coef(fit.mle)[2]
summary(a2_mle)# To calculate the parameters (a)
summary(b2_mle)# To calculate the parameters (b)
confint(fit.mle)# To calculate the confidence interval (a and b ) by MLE
x2.logis <- fitdist(y2, "logis", method="mle")
B<- bootdist(x2.logis, bootmethod="param", niter=1001)
summary(B)
b <- B$estim
a2<-c(b$location)
b2<-c(b$scale)
#To calculate the parameters (a and b) with data sets x3
y.n<-length(y3)
y.location<-mean(y3)
y.var<-(y.n-1)/y.n*var(y3)
y.scale<-sqrt(3*y.var)/pi
library(stats4)
ll.logis<-function(location=y.location,scale=y.scale){-sum(dlogis(y3,location,scale,log=TRUE))}
fit.mle<-mle(ll.logis,method="Nelder-Mead")
a3_mle<-coef(fit.mle)[1]
b3_mle<-coef(fit.mle)[2]
summary(a3_mle)# To calculate the parameters (a)
summary(b3_mle)# To calculate the parameters (b)
confint(fit.mle)# To calculate the confidence interval (a and b ) by MLE
x3.logis <- fitdist(y3, "logis", method="mle")
C <- bootdist(x3.logis, bootmethod="param", niter=1001)
summary(C)
c<- C$estim
a3<-c(c$location)
b3<-c(c$scale)
#To calculate the parameters (a and b) with data sets x4
y.n<-length(y4)
y.location<-mean(y4)
y.var<-(y.n-1)/y.n*var(y4)
y.scale<-sqrt(3*y.var)/pi
library(stats4)
ll.logis<-function(location=y.location,scale=y.scale){-sum(dlogis(y4,location,scale,log=TRUE))}
fit.mle<-mle(ll.logis,method="Nelder-Mead")
a4_mle<-coef(fit.mle)[1]
b4_mle<-coef(fit.mle)[2]
summary(a4_mle)# To calculate the parameters (a)
summary(b4_mle)# To calculate the parameters (b)
confint(fit.mle)# To calculate the confidence interval (a and b ) by MLE
x4.logis <- fitdist(y4, "logis", method="mle")
D <- bootdist(x4.logis, bootmethod="param", niter=1001)
summary(D)
d <- D$estim
a4<-c(d$location)
b4<-c(d$scale)