I have 2x N amount of 1D Signals in files where Column 1 is Signal 1 and Column 2 Signal 2.
Code 1 is simplified example about 1x N amount of 1D signals, while Code 2 is the actual target with two pieces of pseudocode about:
- to create two dimensional vector (
files[[i]] = i,i+1) - just two integer data units in each row separated by comma, and - and then accessing the data there later (
tcrossprod( files[[]][, 2], files[[]][, 2] )) where I cannot refer to all columns 2 of all signals
Simplified Code 1 works as expected
## Example with 1D vector in Single column
N <- 7
files <- vector("list", N)
# Make a list of two column data
for (i in 1:N) {
files[[i]] = i
}
str(files)
# http://stackoverflow.com/a/40323768/54964
tcrossprod( files, files )
Code 2 is pseudocode but target
## Example with 2x1D vectors in two columns
N <- 7
files <- vector("list", N)
# Make a list of two column data
for (i in 1:N) {
files[[i]] = i,i+1 # PSEUDOCODE
}
str(files)
# access one signal single columns by files[[1]][,1] and files[[1]][,2]
tcrossprod( files[[]][, 2], files[[]][, 2] ) # PSEUDOCODE
Assume Vector 1 dimensions are Nx1 and Vector 1 1xM.
Each cell, accessed for instance for Signal 2 Column 2 by files[[1]][,2] contains 1D signal.
Mutiply all such signals of Column 2 by trossprod, you should get the expected result: NxM matrix.
Mathematical description
Data: a list of two columns, where first column is 1D signal; 2nd column is improved 1D signal. I want to compare those improved 1D signals all together in the matrix. Expected output
cor Improved 1 Improved 2 ...
Improved 1 1 0.55
Improved 2 0.111 1
...
I am not tied to any particular R data structures .
Column and cell are just my descriptions of the items in the data units. So not precise because I am newbie in R.
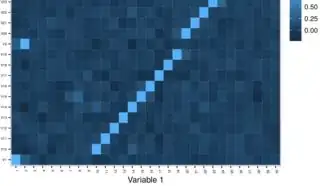
Output of tchakravarty's graphic code in my system where you see x-axis is correct but not y-axis
OS: Debian 8.5
R: 3.1.1