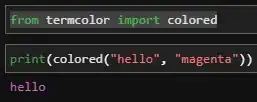
You can output side-by-side figures in an rmarkdown document using the chunk argument fig.show='hold'. Another option is to use ggplot2 to create the plot, instead of the cars scatterplot function. I show both approaches below.
Side-by-Side cars::scatterplot plots in a rmarkdown document
Below is an example with PDF output. out.width='3in' sets the actual size of each plot in the output document, regardless of fig.height and fig.width. But you can still adjust fig.height and fig.width to adjust the aspect ratio and the text size relative to the plot area.
---
title: "Untitled"
author: "eipi10"
date: "November 23, 2016"
output: pdf_document
---
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE)
```
```{r fig.show='hold', out.width='3in', fig.height=5, fig.width=5}
library(car)
scatterplot(Sepal.Length~Sepal.Width|Species,data=iris,grid="FALSE",
boxplots="", reg.line="FALSE", pch=c(0,1,2))
scatterplot(Petal.Width~Sepal.Width|Species,data=iris,grid="FALSE",
boxplots="", reg.line="FALSE", pch=c(0,1,2))
```
Side-by-Side scatterplots using ggplot2
If you're willing to go with ggplot2 then you can get the two-plot layout relatively easily:
library(ggplot2)
library(gridExtra)
theme_set(theme_bw())
grid.arrange(
ggplot(iris, aes(Sepal.Width, Sepal.Length, colour=Species)) +
geom_point() +
geom_smooth(alpha=0.2) +
theme(legend.position="top"),
ggplot(iris, aes(Sepal.Width, Petal.Width, colour=Species)) +
geom_point() +
geom_smooth(alpha=0.2) +
theme(legend.position="top"),
ncol=2)

Customize ggplot2 plots to look more like cars::scatterplot output
You can customize the code above in other ways. If you don't want the confidence bands, add se=FALSE to geom_smooth. If you want different shapes for each species, add aes(shape=Species) to geom_point. If you want the specific shapes used in base graphics, add + scale_shape_manual(values=0:2), etc. You can also get a single legend with a little extra work.
In the code below, I've added these and other customizations to reproduce something closer to your original base graphics plot.
# Components we'll reuse for both plots
my_theme = list(geom_point(aes(shape=Species)),
geom_smooth(se=FALSE, show.legend=FALSE, lwd=0.8),
scale_shape_manual(values=0:2),
scale_colour_manual(values=c("black", "red","green")),
theme_bw(),
theme(panel.grid.major=element_blank(),
panel.grid.minor=element_blank(),
legend.position="top"))
p1 = ggplot(iris, aes(Sepal.Width, Sepal.Length, colour=Species)) +
my_theme +
labs(x="Sepal Width", y="Sepal Length") +
scale_y_continuous(limits=c(3,8)) +
scale_x_continuous(limits=c(1,5))
p2 = ggplot(iris, aes(Sepal.Width, Petal.Width, colour=Species)) +
my_theme +
labs(x="Sepal Width", y="Petal Width")
# Function to extract legend
# https://github.com/hadley/ggplot2/wiki/Share-a-legend-between-two-ggplot2-graphs
g_legend<-function(a.gplot){
tmp <- ggplot_gtable(ggplot_build(a.gplot))
leg <- which(sapply(tmp$grobs, function(x) x$name) == "guide-box")
legend <- tmp$grobs[[leg]]
return(legend)}
leg = g_legend(p1)
grid.arrange(leg,
arrangeGrob(grobs=lapply(list(p1,p2), function(p) p + guides(colour=FALSE, shape=FALSE)), ncol=2),
ncol=1, heights=c(1,10))