How to outline pixel boundaries in matplotlib? For instance, for a semi-random dataset like the one below,
# the code block that follows is irrelevant
import numpy as np
k = []
for s in [2103, 1936, 2247, 2987]:
np.random.seed(s)
k.append(np.random.randint(0, 2, size=(2,6)))
arr = np.hstack([np.vstack(k)[:, :-1], np.vstack(k).T[::-1].T ])
image = np.zeros(shape=(arr.shape[0]+2, arr.shape[1]+2))
image[1:-1, 1:-1] = arr
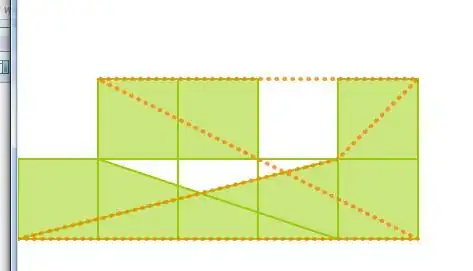
it is quite clear that a contour matching the pixel edges of image would be preferred to the default behavior of the contour function, where the contour lines are effectively drawn across the diagonals of edge pixels.
import matplotlib.pyplot as plt
plt.contour(image[::-1], [0.5], colors='r')
How to make the contours align with the pixels? I'm looking for a solution within numpy and matplotlib libraries.