I am trying to do non-linear regression using R genetic package (rgp) using technique used here: Fitting a curve to specific data (see second method). I am using R package drc for heartrate data:
library(drc)
head(heartrate)
# pressure rate
#1 50.85 348.76
#2 54.92 344.45
#3 59.23 343.05
#4 61.91 332.92
#5 65.22 315.31
#6 67.79 313.50
library(rgp)
res <- symbolicRegression(rate ~ pressure, data=heartrate)
(symbreg <- res$population[[which.min(sapply(res$population, res$fitnessFunction))]])
#function (pressure)
#pressure + (pressure/0.853106872646055 + pressure)
ggplot() +
geom_point(data=heartrate, aes(pressure,rate), size = 3) +
geom_line(data=data.frame(symbx=heartrate$pressure,
symby=sapply(heartrate$pressure, symbreg)),
aes(symbx, symby), colour = "red")
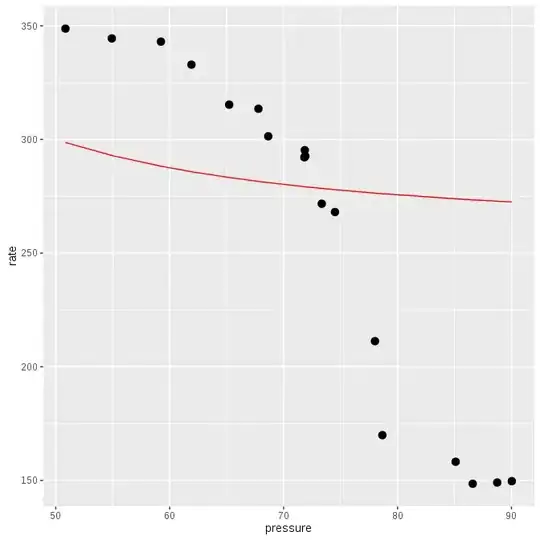
However, the resulting regression line that I am getting is clearly incorrect. The distribution of data points indicate a curvilinear relation with rate reducing as pressure increases (inverse association). However, the regression line generated is linear and in the wrong direction.
Where is the error?
Edit:
Using increased steps as suggested by @cuttlefish44 in comments:
res = symbolicRegression(rate ~ pressure, data = heartrate, stopCondition = makeStepsStopCondition(45000))
(symbreg <- res$population[[which.min(sapply(res$population, res$fitnessFunction))]])
#function (pressure)
#exp(exp(exp(cos(cos(-9.23878724686801/pressure)))))
It took 8 minutes to complete. The plot is:
The direction of regression line is better than above (!), but it indicates that it will take a really long time to reach the obvious direction. The regression line with the function obtained by @cuttlefish44 is also similar and not a really good fit.