I am trying to compute the Mahalanobis distance between each observations of a dataset dat, where each row is an observation and each column is a variable. Such distance is defined as:
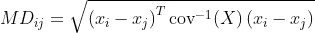
I wrote a function that does it, but I feel like it is slow. Is there any better way to compute this in R ?
To generate some data to test the function:
generateData <- function(nObs, nVar){
library(MASS)
mvrnorm(n=nObs, rep(0,nVar), diag(nVar))
}
This is the function I have written so far. They both work and for my data (800 obs and 90 variables), it takes approximatively 30 and 33 seconds for the method = "forLoop" and method = "apply", respectively.
mhbd_calc2 <- function(dat, method) { #Method is either "forLoop" or "apply"
dat <- as.matrix(na.omit(dat))
nObs <- nrow(dat)
mhbd <- matrix(nrow=nObs,ncol = nObs)
cv_mat_inv = solve(var(dat))
distMH = function(x){ #Mahalanobis distance function
diff = dat[x[1],]-dat[x[2],]
diff %*% cv_mat_inv %*% diff
}
if(method=="forLoop")
{
for (i in 1:nObs){
for(j in 1:i){
mhbd[i,j] <- distMH(c(i,j))
}
}
}
if(method=="apply")
{
mhbd[lower.tri(mhbd)] = apply(combn(nrow(dat),2),2, distMH)
}
result = sqrt(mhbd)
colnames(result)=rownames(dat)
rownames(result)=rownames(dat)
return(as.dist(result))
}
NB: I tried using outer() but it was even slower (60seconds)