igraph
We start by making a graph from your adjacency matrix:
df <- t(df)
ga <- graph.adjacency(as.matrix(df), weighted = TRUE, mode = "directed")
Then, plot a circle:
par(mar = rep(0.25, 4))
pts <- seq(0, 2*pi, l = 100)
plot(cbind(sin(pts), cos(pts)), type = "l", frame = F, xaxt = "n", yaxt = "n")
Finally, plot the graph:
plot.igraph(ga,
vertex.label = V(ga)$name,
edge.width = E(ga)$weight,
edge.curved = TRUE,
edge.label = E(ga)$weight,
layout = layout_in_circle(ga, order = V(ga)),
add = T)
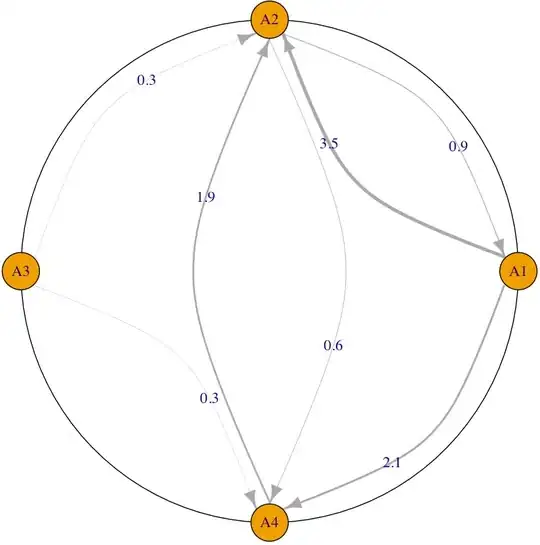
Output below. You can customize your graph (e.g. curvature and colors of edges, shapes of vertices) as desired.
ggplot2
The main idea is to set up three sets of geoms: the circle, the nodes (vertices), and the lines (edges). First, we load some packages, and prep the circle and nodes:
library(ggplot2)
library(tidyr)
library(dplyr)
# For circle
pts <- seq(0, 2*pi, l = 100)
# For nodes
theta <- seq(0, 2*pi, l = nrow(df) + 1)[1:nrow(df)]
l <- data.frame(x = sin(theta), y = cos(theta), v = names(df),
stringsAsFactors = FALSE)
The edges are a little bit more involved. I make a function to make coordinates for the lines, given an origin and destination:
make_edge <- function(origin, dest, l, shrink = .9) {
# l is the layout matrix for the nodes that we made previously
data.frame(
x0 = l$x[l$v == origin],
y0 = l$y[l$v == origin],
x1 = l$x[l$v == dest],
y1 = l$y[l$v == dest]
) * shrink
}
Then, we make an adjacency graph, and bind the edge coordinates to it:
gr <- gather(mutate(df, dest = names(df)), origin, wt, -dest)
gr <- gr[gr$wt != 0, ]
edges <- do.call(rbind,
mapply(make_edge, gr$origin, gr$dest, list(l), shrink = .94, SIMPLIFY = F)
)
ga <- cbind(gr, edges)
Finally, we plot:
ggplot() +
geom_path(data = data.frame(x = sin(pts), y = cos(pts)), aes(x, y)) +
geom_label(data = l, aes(x, y, label = v)) +
geom_curve(data = ga,
aes(x = x0, y = y0, xend = x1, yend = y1, size = wt, colour = origin),
alpha = 0.8,
curvature = 0.1,
arrow = arrow(length = unit(2, "mm"))) +
scale_size_continuous(range=c(.25,2), guide = FALSE) +
theme_void()
Output: