Without using fixed thresholds, you can try to cluster the image by gray levels. As a preprocessing step, I would suggest using morphological opening to make the difference of gray levels of the neighboring pixels small, so you have less noise in the clustered image.
Below I apply two successive morphological openings to the image using a 3x3 circular kernel, and then apply k-means clustering to the gray levels. From your sample images and some I found on the internet, I decided to set k = 4. If you are using high-resolution images, first downsample them to dimensions ~400-600. Otherwise the morphological operation may not have a significant effect, and the k-means will be slow.
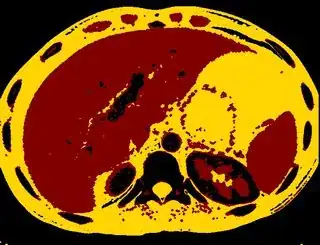
Below are some of the opened and segmented images. Of course there's more to be done in terms of
- separating out the liver region
- generalizing this to a large dataset
but hope this is at least a starting point.
I don't have matlab, so the code is in c++ and opencv, but the conversion should be simple as it involves only morphological and clustering operations, it should be somewhat similar to this.
Update
You might be able to narrow down the region or interest by filtering out the darkest and the lightest regions from the segmented image. For this, use the k-means cluster centers, check for the extreme values (max and min) and remove the corresponding k values from the labeled image. Then you can look for large structures to the left of the result image. Worst case, you might get a hole on the left side when the extreme region filtering goes wrong. I've updated the code and results.
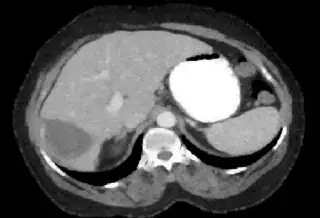
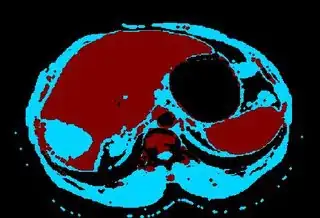
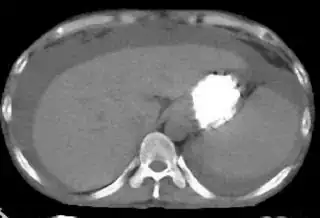
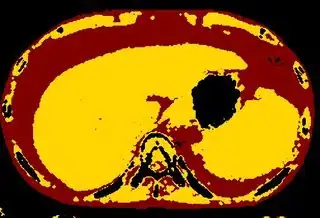
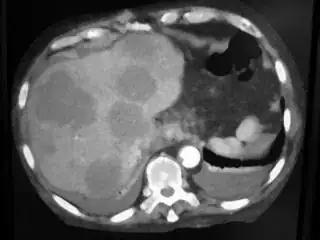
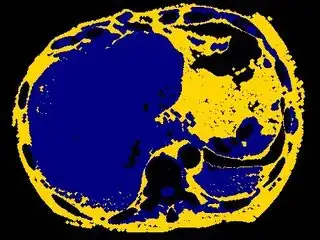
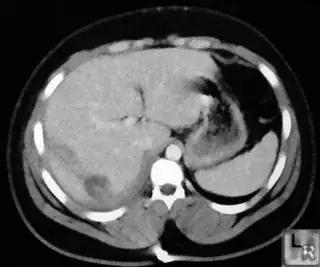
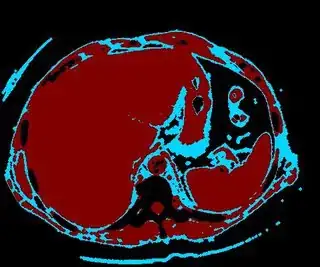


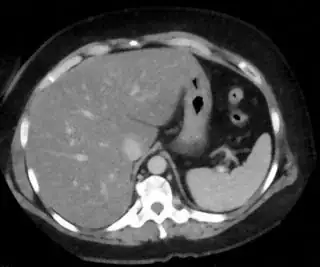
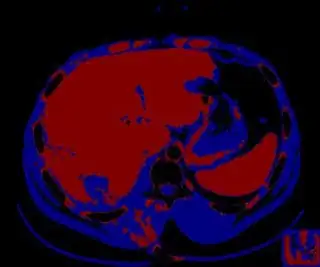
opencv c++ code
// load image as gray scale
Mat im = imread("5.jpg", 0);
// morphological opening
Mat kernel = getStructuringElement(MORPH_ELLIPSE, Size(3, 3));
Mat morph;
morphologyEx(im, morph, CV_MOP_OPEN, kernel, Point(-1, -1), 2);
// clustering
int k = 4;
Mat segment, lbl;
vector<float> centers;
morph.convertTo(segment, CV_32F);
int imsize[] = {segment.rows, segment.cols};
Mat color = segment.reshape(1, segment.rows*segment.cols);
kmeans(color, k, lbl, TermCriteria(CV_TERMCRIT_EPS+CV_TERMCRIT_ITER, 10, 1.0), k, KMEANS_PP_CENTERS, centers);
lbl = lbl.reshape(1, 2, imsize);
// find argmin and argmax to find extreme gray level regions
int minidx = min_element(centers.begin(), centers.end()) - centers.begin();
int maxidx = max_element(centers.begin(), centers.end()) - centers.begin();
// prepare a mask to filter extreme gray level regions
Mat mask = (lbl != minidx) ^ (lbl == maxidx);
// only for display purposes
Mat lbldisp;
lbl.convertTo(lbldisp, CV_8U, 255.0/(k-1));
Mat lblColor;
applyColorMap(lbldisp, lblColor, COLORMAP_JET);
// region of interest
Mat roiColor = Mat::zeros(lblColor.size(), CV_8UC3);
lblColor.copyTo(roiColor, mask);
imshow("opened", morph);
imshow("segmented", lblColor);
imshow("roi", roiColor);
waitKey();
 .
.  .
My implementation code is
.
My implementation code is  .
.